Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
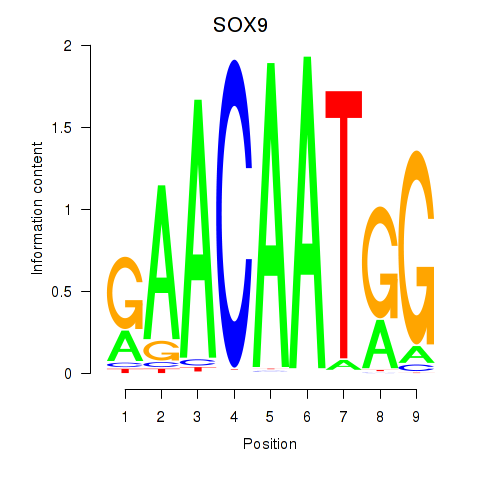
Results for SOX9
Z-value: 0.88
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.18 | 3.9e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_102106829 | 2.25 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr7_+_77167343 | 2.14 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr4_+_41614720 | 1.99 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_-_11871815 | 1.96 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr17_-_42276574 | 1.90 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr4_+_41614909 | 1.89 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_-_51027838 | 1.86 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_-_149051194 | 1.85 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr7_+_155090271 | 1.66 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr17_-_14683517 | 1.60 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr1_-_85930246 | 1.53 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr3_-_149051444 | 1.36 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr6_-_75915757 | 1.33 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr5_-_43313574 | 1.32 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chrX_-_117107542 | 1.21 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr12_-_6233828 | 1.16 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr14_+_53019993 | 1.13 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr19_+_10765699 | 1.13 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr1_+_164528866 | 1.12 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr11_-_10830463 | 1.08 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr4_-_186732048 | 1.03 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_74633036 | 1.00 |
ENST00000343975.5
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chrX_+_84499081 | 0.95 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr12_-_25055177 | 0.95 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_88974236 | 0.94 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chrX_-_45060135 | 0.93 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr8_-_80993010 | 0.93 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr20_-_39317868 | 0.93 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr8_+_70378852 | 0.91 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr1_+_185703513 | 0.90 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr5_+_74632993 | 0.89 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr14_-_38725573 | 0.85 |
ENST00000342213.2
|
CLEC14A
|
C-type lectin domain family 14, member A |
| chr14_+_63671105 | 0.85 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr1_+_29241027 | 0.80 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr14_-_65569057 | 0.80 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr6_+_37787704 | 0.79 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr11_+_63953691 | 0.79 |
ENST00000543847.1
|
STIP1
|
stress-induced-phosphoprotein 1 |
| chr9_-_139658965 | 0.79 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr11_+_128563948 | 0.75 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_-_70475701 | 0.71 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr21_+_26934165 | 0.71 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr1_-_182360498 | 0.71 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr17_+_26646175 | 0.71 |
ENST00000583381.1
ENST00000582113.1 ENST00000582384.1 |
TMEM97
|
transmembrane protein 97 |
| chr8_-_38326119 | 0.70 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr3_-_112329110 | 0.70 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chrX_+_41193407 | 0.70 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr7_+_16793160 | 0.69 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chrX_+_48380205 | 0.68 |
ENST00000446158.1
ENST00000414061.1 |
EBP
|
emopamil binding protein (sterol isomerase) |
| chr1_-_173991434 | 0.68 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr14_-_88459182 | 0.68 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr4_+_78079450 | 0.67 |
ENST00000395640.1
ENST00000512918.1 |
CCNG2
|
cyclin G2 |
| chr4_-_110723194 | 0.66 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr14_+_32546145 | 0.65 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr19_+_16187816 | 0.65 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr9_-_123476612 | 0.65 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr15_-_59041768 | 0.65 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr1_-_68698197 | 0.64 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr8_-_82395461 | 0.63 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr9_-_123476719 | 0.63 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr2_+_111880242 | 0.63 |
ENST00000393252.3
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr4_+_78079570 | 0.63 |
ENST00000509972.1
|
CCNG2
|
cyclin G2 |
| chr2_-_208031542 | 0.62 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_65583561 | 0.62 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr7_+_128379449 | 0.62 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr2_-_70475730 | 0.62 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr20_-_17662705 | 0.61 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr7_-_75368248 | 0.61 |
ENST00000434438.2
ENST00000336926.6 |
HIP1
|
huntingtin interacting protein 1 |
| chr6_-_11232891 | 0.61 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr18_-_53177984 | 0.61 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr18_+_6729698 | 0.60 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr6_+_89790459 | 0.60 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_+_11660227 | 0.59 |
ENST00000443614.2
ENST00000525900.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chrX_+_84498989 | 0.59 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr4_+_54243917 | 0.59 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr8_-_38325219 | 0.57 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr17_-_76713100 | 0.57 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr18_+_42260861 | 0.57 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr7_+_77167376 | 0.56 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chrX_+_84499038 | 0.56 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr12_+_110011571 | 0.55 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr5_-_131826457 | 0.55 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr3_+_25469802 | 0.55 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr4_-_110723335 | 0.54 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr5_+_140797296 | 0.52 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr2_+_54683419 | 0.52 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_-_39425222 | 0.52 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_110723134 | 0.51 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr17_-_39728303 | 0.51 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chrX_+_41192595 | 0.51 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr12_-_110011288 | 0.50 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr11_+_128563652 | 0.50 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_140602904 | 0.49 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr19_+_8455200 | 0.48 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr7_-_22233442 | 0.48 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr20_-_17662878 | 0.48 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr14_-_53019211 | 0.47 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr2_-_70475586 | 0.47 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_+_217498105 | 0.47 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chrX_-_2418596 | 0.47 |
ENST00000381218.3
|
ZBED1
|
zinc finger, BED-type containing 1 |
| chr2_+_109271481 | 0.47 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_+_11660120 | 0.46 |
ENST00000220584.4
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_+_208576259 | 0.46 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr2_-_175629135 | 0.46 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr5_-_39425068 | 0.46 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr9_-_74979420 | 0.46 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr9_+_116263639 | 0.46 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chrX_+_135579238 | 0.45 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr11_-_126138808 | 0.45 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr3_+_44690211 | 0.44 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr1_+_66458072 | 0.44 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr5_-_39425290 | 0.44 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr11_-_77531752 | 0.44 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr15_-_52944231 | 0.44 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr3_+_25469724 | 0.44 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr6_+_89790490 | 0.44 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr1_-_93645818 | 0.43 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr14_-_88459503 | 0.43 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr1_+_12834984 | 0.43 |
ENST00000357726.4
|
PRAMEF12
|
PRAME family member 12 |
| chr9_+_116263778 | 0.43 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr15_-_50411412 | 0.42 |
ENST00000284509.6
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr4_+_71570430 | 0.42 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_+_8378140 | 0.41 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr19_-_10491130 | 0.41 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr12_-_71003568 | 0.41 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_-_88108212 | 0.41 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr5_-_141338627 | 0.40 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr5_+_138677515 | 0.40 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr12_-_31479107 | 0.40 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr7_+_116312411 | 0.40 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr22_-_19512893 | 0.39 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr19_+_48972459 | 0.39 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr16_+_2083265 | 0.38 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr8_-_38326139 | 0.38 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr19_+_8455077 | 0.38 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr4_-_175204765 | 0.38 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr10_+_6244829 | 0.38 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr15_+_84115868 | 0.38 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr15_+_85923797 | 0.37 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_10829851 | 0.37 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_24286287 | 0.37 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_+_156119466 | 0.37 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr16_+_3068393 | 0.36 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr7_+_134464376 | 0.36 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr16_+_3704822 | 0.35 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr10_-_81205373 | 0.35 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr5_+_140739537 | 0.35 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr1_-_153643442 | 0.35 |
ENST00000368681.1
ENST00000361891.4 |
ILF2
|
interleukin enhancer binding factor 2 |
| chr11_+_63953587 | 0.34 |
ENST00000305218.4
ENST00000538945.1 |
STIP1
|
stress-induced-phosphoprotein 1 |
| chr7_+_128379346 | 0.34 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr10_+_97803151 | 0.34 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr5_+_140729649 | 0.34 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr4_+_113152978 | 0.34 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr4_-_2264015 | 0.33 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr4_+_154125565 | 0.33 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr4_+_54243798 | 0.33 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr14_-_71276211 | 0.33 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr7_+_128349106 | 0.33 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr2_+_54198210 | 0.33 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr4_+_78078304 | 0.33 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr1_+_33722080 | 0.32 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr9_-_99064386 | 0.32 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr1_-_21503337 | 0.32 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr16_-_25026641 | 0.32 |
ENST00000289968.6
ENST00000303665.5 ENST00000455311.2 ENST00000441763.2 |
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr12_+_49761147 | 0.32 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr1_-_182641367 | 0.32 |
ENST00000508450.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr14_+_102276192 | 0.31 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_+_26646121 | 0.31 |
ENST00000226230.6
|
TMEM97
|
transmembrane protein 97 |
| chr19_+_45504688 | 0.31 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_+_113152881 | 0.31 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_175629164 | 0.31 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_228337553 | 0.31 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr9_-_72287191 | 0.31 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr18_-_44497308 | 0.31 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chrX_+_129473859 | 0.30 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr5_+_140593509 | 0.30 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr14_-_91884150 | 0.30 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr12_-_49075941 | 0.30 |
ENST00000553086.1
ENST00000548304.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr1_-_119530428 | 0.30 |
ENST00000369429.3
|
TBX15
|
T-box 15 |
| chr9_-_73483958 | 0.30 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_91546926 | 0.29 |
ENST00000550758.1
|
DCN
|
decorin |
| chr2_-_240322643 | 0.29 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr3_-_101232019 | 0.29 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr14_-_91884115 | 0.29 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr18_-_34408902 | 0.29 |
ENST00000593035.1
ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr19_+_13906250 | 0.29 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr15_-_34659349 | 0.29 |
ENST00000314891.6
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr1_+_33116765 | 0.29 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chrX_+_70503037 | 0.29 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr12_+_69979210 | 0.29 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr19_-_44100275 | 0.29 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr10_+_69644404 | 0.29 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr2_+_170590321 | 0.28 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr7_+_28452130 | 0.28 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr12_+_53443963 | 0.28 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr14_-_73493784 | 0.28 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr1_+_25071848 | 0.28 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr11_-_69519410 | 0.28 |
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19 |
| chr11_+_34073269 | 0.27 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr17_+_67498538 | 0.27 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_140557371 | 0.27 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.6 | 1.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.6 | 1.7 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 | 1.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 0.9 | GO:0070668 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 0.9 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 0.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 3.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 0.7 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.2 | 0.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.6 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.2 | 0.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 1.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 0.6 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 3.3 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.2 | 1.0 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.5 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.7 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.2 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 1.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.5 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 1.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.9 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.9 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.8 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.2 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 1.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 1.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:2000584 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 1.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.0 | GO:0035787 | cell migration involved in kidney development(GO:0035787) |
| 0.0 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 1.7 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.5 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.9 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 1.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.2 | 1.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 1.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 3.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 0.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 2.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.6 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 1.0 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.5 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 1.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 1.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.4 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.2 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 1.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035473 | high-density lipoprotein particle binding(GO:0008035) lipase binding(GO:0035473) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.7 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |