Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
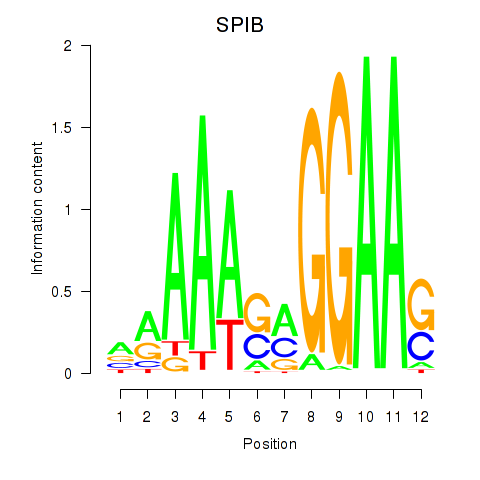
Results for SPIB
Z-value: 0.64
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | -0.25 | 2.3e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_13213662 | 0.88 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr3_-_116163830 | 0.80 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_-_29641084 | 0.78 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr6_+_136172820 | 0.74 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr19_+_50919056 | 0.74 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr1_+_120839412 | 0.72 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr17_-_72709050 | 0.71 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr1_+_206138884 | 0.70 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr10_-_121302195 | 0.67 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr8_+_125985531 | 0.55 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr5_-_66492562 | 0.55 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr13_+_108922228 | 0.53 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr13_+_31309645 | 0.50 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr16_-_4850471 | 0.49 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr2_+_96991935 | 0.49 |
ENST00000361124.4
ENST00000420728.1 ENST00000542887.1 |
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr11_-_64570706 | 0.47 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr16_+_54964740 | 0.47 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr9_+_109625378 | 0.44 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr5_+_35856951 | 0.44 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr7_-_36764004 | 0.42 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr3_+_125694347 | 0.40 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr2_-_231084820 | 0.40 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr1_-_31230650 | 0.39 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr2_-_100759037 | 0.38 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr16_+_30960375 | 0.38 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr10_-_74856608 | 0.38 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr3_-_121379739 | 0.37 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr7_-_36764142 | 0.37 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_-_143913143 | 0.37 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr1_+_120839005 | 0.37 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr1_+_206138457 | 0.36 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr12_-_9885888 | 0.35 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr20_+_62492566 | 0.35 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr2_-_242089677 | 0.35 |
ENST00000405260.1
|
PASK
|
PAS domain containing serine/threonine kinase |
| chr19_-_17375527 | 0.35 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr8_-_77912431 | 0.34 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr9_-_130541017 | 0.33 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr19_+_42381337 | 0.33 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr12_+_50355647 | 0.33 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr6_+_49431073 | 0.33 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr17_+_7461613 | 0.33 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_43019698 | 0.33 |
ENST00000431905.1
ENST00000294973.6 |
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr6_-_139308777 | 0.33 |
ENST00000529597.1
ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr1_+_43855545 | 0.32 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr5_-_64858944 | 0.32 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr10_+_49917751 | 0.32 |
ENST00000325239.5
ENST00000413659.2 |
WDFY4
|
WDFY family member 4 |
| chr9_-_95640218 | 0.32 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr9_-_34397800 | 0.31 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr10_-_79789291 | 0.31 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr11_-_64851496 | 0.29 |
ENST00000404147.3
ENST00000275517.3 |
CDCA5
|
cell division cycle associated 5 |
| chr19_-_48867171 | 0.29 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr16_+_30194916 | 0.29 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr19_-_48867291 | 0.28 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr11_+_131240373 | 0.27 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chrX_+_109246285 | 0.27 |
ENST00000372073.1
ENST00000372068.2 ENST00000288381.4 |
TMEM164
|
transmembrane protein 164 |
| chr11_+_60467047 | 0.27 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr2_-_225266711 | 0.27 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr1_-_111506562 | 0.27 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr1_-_120935894 | 0.27 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr19_+_14551066 | 0.26 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr15_-_33360342 | 0.26 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr2_-_225266743 | 0.26 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr8_-_125551278 | 0.26 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr14_+_24616588 | 0.26 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr4_+_95129061 | 0.26 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_-_87281196 | 0.26 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_-_190044480 | 0.26 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_87281224 | 0.25 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_131148538 | 0.25 |
ENST00000541421.2
|
SMLR1
|
small leucine-rich protein 1 |
| chr3_+_29322437 | 0.25 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr22_-_37640277 | 0.25 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_+_159272111 | 0.25 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr19_+_35940486 | 0.25 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr5_-_58335281 | 0.25 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_-_231084617 | 0.25 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr9_+_120466650 | 0.25 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chr8_+_134203303 | 0.25 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr8_+_74903580 | 0.24 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr9_+_104296122 | 0.24 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr7_+_106505696 | 0.24 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chrX_-_153637612 | 0.24 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr3_+_16926441 | 0.24 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr5_+_159848854 | 0.23 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr2_+_113885138 | 0.23 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr1_-_145039949 | 0.23 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_-_122240792 | 0.23 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr14_-_98444438 | 0.23 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr1_-_153919128 | 0.23 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr16_-_50715196 | 0.23 |
ENST00000423026.2
|
SNX20
|
sorting nexin 20 |
| chr19_+_2476116 | 0.23 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr10_-_62149433 | 0.22 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_202125219 | 0.22 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_+_173940163 | 0.22 |
ENST00000539448.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr11_+_46722368 | 0.22 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr17_-_5138099 | 0.22 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chrX_-_47489244 | 0.22 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr4_-_80994619 | 0.22 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr15_+_50716576 | 0.22 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr11_+_64107663 | 0.22 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr2_+_127413677 | 0.21 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr16_-_73082274 | 0.21 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr11_-_47399942 | 0.21 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_+_68592305 | 0.21 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr11_-_47400078 | 0.21 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr10_-_52008313 | 0.21 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr7_-_121944491 | 0.20 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr2_+_127413704 | 0.20 |
ENST00000409836.3
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr6_-_139309378 | 0.20 |
ENST00000450536.2
ENST00000258062.5 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr1_+_209941827 | 0.20 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr4_-_85887503 | 0.20 |
ENST00000509172.1
ENST00000322366.6 ENST00000295888.4 ENST00000502713.1 |
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr8_-_101734170 | 0.20 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_169064245 | 0.20 |
ENST00000256935.8
|
DOCK2
|
dedicator of cytokinesis 2 |
| chr3_+_46395219 | 0.19 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr19_-_17375541 | 0.19 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr2_-_26101374 | 0.19 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr1_+_161475208 | 0.19 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr6_+_36973406 | 0.19 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr13_-_111214015 | 0.19 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr7_+_106505912 | 0.19 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr11_+_47290914 | 0.19 |
ENST00000342922.4
|
MADD
|
MAP-kinase activating death domain |
| chr1_+_151584544 | 0.19 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr11_+_1860832 | 0.18 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr7_+_141811539 | 0.18 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chrX_+_24072833 | 0.18 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr15_+_42651691 | 0.18 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr16_+_57576584 | 0.18 |
ENST00000340339.4
|
GPR114
|
G protein-coupled receptor 114 |
| chr6_-_49430886 | 0.18 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr9_-_99145957 | 0.18 |
ENST00000375257.1
ENST00000253270.7 ENST00000375259.4 |
SLC35D2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr2_-_113993020 | 0.18 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr8_-_101734308 | 0.18 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr22_-_37915535 | 0.18 |
ENST00000403299.1
|
CARD10
|
caspase recruitment domain family, member 10 |
| chr19_+_49838653 | 0.18 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr1_-_20446020 | 0.18 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr2_-_152118352 | 0.18 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr2_+_143886877 | 0.18 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr15_-_33360085 | 0.18 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chr3_-_178984759 | 0.18 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr1_+_167298281 | 0.18 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr9_+_120466610 | 0.18 |
ENST00000394487.4
|
TLR4
|
toll-like receptor 4 |
| chr13_+_53602894 | 0.18 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr6_-_154677900 | 0.17 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr11_-_47400062 | 0.17 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr10_+_17851362 | 0.17 |
ENST00000331429.2
ENST00000457317.1 |
MRC1L1
|
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr14_+_85996471 | 0.17 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_-_14029515 | 0.17 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr5_+_40909354 | 0.17 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr2_-_231084659 | 0.17 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr7_-_99756293 | 0.17 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr7_+_86781847 | 0.17 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr16_+_57392684 | 0.17 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr2_+_233925064 | 0.16 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr10_+_11206925 | 0.16 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_+_109237717 | 0.16 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_39093672 | 0.16 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr10_+_114133773 | 0.16 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr19_-_3786253 | 0.16 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr11_+_1861399 | 0.16 |
ENST00000381905.3
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr22_-_37640456 | 0.16 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr8_+_121457642 | 0.16 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chrX_-_74376108 | 0.16 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr4_-_140005443 | 0.16 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr19_-_3786354 | 0.16 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_-_150038704 | 0.16 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr14_+_102228123 | 0.16 |
ENST00000422945.2
ENST00000554442.1 ENST00000556260.2 ENST00000328724.5 ENST00000557268.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr22_-_40859415 | 0.15 |
ENST00000402630.1
ENST00000407029.1 |
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_+_85996507 | 0.15 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr12_+_49717081 | 0.15 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr11_-_8680383 | 0.15 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr16_-_30537839 | 0.15 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr7_+_86781677 | 0.15 |
ENST00000331242.7
ENST00000394702.3 ENST00000413276.2 ENST00000446796.2 ENST00000411766.2 ENST00000420131.1 ENST00000414630.2 ENST00000453049.1 ENST00000428819.1 ENST00000448598.1 ENST00000449088.3 ENST00000430405.3 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr11_+_61100676 | 0.15 |
ENST00000394900.3
ENST00000532173.2 |
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae) |
| chr20_+_2821340 | 0.15 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr2_+_85981008 | 0.15 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr17_+_19030782 | 0.15 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr14_+_45431379 | 0.15 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr3_-_52273098 | 0.15 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr1_+_161632937 | 0.15 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr20_-_20033052 | 0.15 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr7_+_139528952 | 0.14 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr2_+_242089833 | 0.14 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr1_+_40840320 | 0.14 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr15_+_91427642 | 0.14 |
ENST00000328850.3
ENST00000414248.2 |
FES
|
feline sarcoma oncogene |
| chr6_+_130339710 | 0.14 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr11_+_22696314 | 0.14 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr19_+_48828788 | 0.14 |
ENST00000594198.1
ENST00000597279.1 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr1_+_104068312 | 0.14 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr14_+_74417192 | 0.14 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr2_-_180871780 | 0.14 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr10_+_18429671 | 0.14 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_-_43855444 | 0.14 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr1_+_46713404 | 0.14 |
ENST00000371975.4
ENST00000469835.1 |
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr6_-_34524093 | 0.14 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr19_-_42192096 | 0.14 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr1_+_46713357 | 0.14 |
ENST00000442598.1
|
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr3_+_46283863 | 0.14 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chrX_-_77582980 | 0.13 |
ENST00000373304.3
|
CYSLTR1
|
cysteinyl leukotriene receptor 1 |
| chr12_-_15114191 | 0.13 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_145039835 | 0.13 |
ENST00000533259.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_+_42952237 | 0.13 |
ENST00000485511.1
ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta |
| chr19_-_42192189 | 0.13 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.2 | 0.2 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.2 | 1.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.6 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.5 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.1 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.4 | GO:0070092 | negative regulation of glycogen biosynthetic process(GO:0045719) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.3 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.3 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0038189 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.4 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 0.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |