|
chr5_+_137801160
Show fit
|
23.15 |
ENST00000239938.4
|
EGR1
|
early growth response 1
|
|
chr10_-_64576105
Show fit
|
12.75 |
ENST00000242480.3
ENST00000411732.1
|
EGR2
|
early growth response 2
|
|
chr8_-_22550815
Show fit
|
6.89 |
ENST00000317216.2
|
EGR3
|
early growth response 3
|
|
chr19_+_45971246
Show fit
|
5.63 |
ENST00000585836.1
ENST00000417353.2
ENST00000353609.3
ENST00000591858.1
ENST00000443841.2
ENST00000590335.1
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr4_+_87928140
Show fit
|
5.48 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr19_+_2476116
Show fit
|
3.80 |
ENST00000215631.4
ENST00000587345.1
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr12_+_52445191
Show fit
|
3.79 |
ENST00000243050.1
ENST00000394825.1
ENST00000550763.1
ENST00000394824.2
ENST00000548232.1
ENST00000562373.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr2_-_106054952
Show fit
|
3.22 |
ENST00000336660.5
ENST00000393352.3
ENST00000607522.1
|
FHL2
|
four and a half LIM domains 2
|
|
chr10_+_112257596
Show fit
|
3.20 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr2_-_106015527
Show fit
|
3.18 |
ENST00000344213.4
ENST00000358129.4
|
FHL2
|
four and a half LIM domains 2
|
|
chr2_-_106015491
Show fit
|
3.16 |
ENST00000408995.1
ENST00000393353.3
ENST00000322142.8
|
FHL2
|
four and a half LIM domains 2
|
|
chr7_-_27183263
Show fit
|
3.06 |
ENST00000222726.3
|
HOXA5
|
homeobox A5
|
|
chr14_+_75745477
Show fit
|
2.57 |
ENST00000303562.4
ENST00000554617.1
ENST00000554212.1
ENST00000535987.1
ENST00000555242.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr2_-_73520667
Show fit
|
2.44 |
ENST00000545030.1
ENST00000436467.2
|
EGR4
|
early growth response 4
|
|
chr6_-_16761678
Show fit
|
2.36 |
ENST00000244769.4
ENST00000436367.1
|
ATXN1
|
ataxin 1
|
|
chr19_-_17356697
Show fit
|
1.99 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr17_+_42634844
Show fit
|
1.80 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2
|
|
chr14_-_74226961
Show fit
|
1.79 |
ENST00000286523.5
ENST00000435371.1
|
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1
|
|
chr1_-_208084729
Show fit
|
1.55 |
ENST00000310833.7
ENST00000356522.4
|
CD34
|
CD34 molecule
|
|
chr2_-_209118974
Show fit
|
1.54 |
ENST00000415913.1
ENST00000415282.1
ENST00000446179.1
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble
|
|
chr3_+_49027308
Show fit
|
1.50 |
ENST00000383729.4
ENST00000343546.4
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr19_+_1026566
Show fit
|
1.49 |
ENST00000348419.3
ENST00000565096.2
ENST00000562958.2
ENST00000562075.2
ENST00000607102.1
|
CNN2
|
calponin 2
|
|
chr19_+_1026298
Show fit
|
1.48 |
ENST00000263097.4
|
CNN2
|
calponin 2
|
|
chr9_+_131644781
Show fit
|
1.39 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr15_+_83776324
Show fit
|
1.35 |
ENST00000379390.6
ENST00000379386.4
ENST00000565774.1
ENST00000565982.1
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr5_-_111093167
Show fit
|
1.28 |
ENST00000446294.2
ENST00000419114.2
|
NREP
|
neuronal regeneration related protein
|
|
chr7_+_5919458
Show fit
|
1.24 |
ENST00000416608.1
|
OCM
|
oncomodulin
|
|
chr9_-_98269481
Show fit
|
1.18 |
ENST00000418258.1
ENST00000553011.1
ENST00000551845.1
|
PTCH1
|
patched 1
|
|
chr3_+_143690640
Show fit
|
1.09 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr17_+_3379284
Show fit
|
1.08 |
ENST00000263080.2
|
ASPA
|
aspartoacylase
|
|
chr9_-_79520989
Show fit
|
1.07 |
ENST00000376713.3
ENST00000376718.3
ENST00000428286.1
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr15_+_96873921
Show fit
|
1.05 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_-_111804905
Show fit
|
1.03 |
ENST00000358835.3
ENST00000435970.1
|
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit
|
|
chr11_-_57282349
Show fit
|
0.95 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1
|
|
chr2_+_87769459
Show fit
|
0.94 |
ENST00000414030.1
ENST00000437561.1
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr4_+_81118647
Show fit
|
0.93 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8
|
|
chr2_-_112237835
Show fit
|
0.92 |
ENST00000442293.1
ENST00000439494.1
|
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding)
|
|
chr8_+_1993152
Show fit
|
0.86 |
ENST00000262113.4
|
MYOM2
|
myomesin 2
|
|
chr5_-_53606396
Show fit
|
0.84 |
ENST00000504924.1
ENST00000507646.2
ENST00000502271.1
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr2_+_191792376
Show fit
|
0.82 |
ENST00000409428.1
ENST00000409215.1
|
GLS
|
glutaminase
|
|
chr9_+_124088860
Show fit
|
0.80 |
ENST00000373806.1
|
GSN
|
gelsolin
|
|
chr1_+_16085244
Show fit
|
0.79 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr12_-_6772303
Show fit
|
0.77 |
ENST00000396807.4
ENST00000446105.2
ENST00000341550.4
|
ING4
|
inhibitor of growth family, member 4
|
|
chr1_-_91487770
Show fit
|
0.77 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644
|
|
chr7_-_44180673
Show fit
|
0.74 |
ENST00000457314.1
ENST00000447951.1
ENST00000431007.1
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr8_+_1993173
Show fit
|
0.72 |
ENST00000523438.1
|
MYOM2
|
myomesin 2
|
|
chr4_+_87928413
Show fit
|
0.72 |
ENST00000544085.1
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr8_+_97597148
Show fit
|
0.68 |
ENST00000521590.1
|
SDC2
|
syndecan 2
|
|
chr6_+_151561085
Show fit
|
0.66 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_176923846
Show fit
|
0.66 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr5_-_176923803
Show fit
|
0.66 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr20_+_35169885
Show fit
|
0.66 |
ENST00000279022.2
ENST00000346786.2
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr2_-_172087824
Show fit
|
0.65 |
ENST00000521943.1
|
TLK1
|
tousled-like kinase 1
|
|
chr12_-_15104040
Show fit
|
0.61 |
ENST00000541644.1
ENST00000545895.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr7_+_94285637
Show fit
|
0.61 |
ENST00000482108.1
ENST00000488574.1
|
PEG10
|
paternally expressed 10
|
|
chr5_-_33892046
Show fit
|
0.57 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr1_+_223889285
Show fit
|
0.54 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr2_-_128432639
Show fit
|
0.54 |
ENST00000545738.2
ENST00000355119.4
ENST00000409808.2
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr15_+_74218787
Show fit
|
0.52 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr19_+_13134772
Show fit
|
0.49 |
ENST00000587760.1
ENST00000585575.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_+_59989791
Show fit
|
0.49 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7
|
|
chr19_-_47975417
Show fit
|
0.48 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr6_-_159466042
Show fit
|
0.48 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr3_+_9834758
Show fit
|
0.47 |
ENST00000485273.1
ENST00000433034.1
ENST00000397256.1
|
ARPC4
ARPC4-TTLL3
|
actin related protein 2/3 complex, subunit 4, 20kDa
ARPC4-TTLL3 readthrough
|
|
chr7_-_107643674
Show fit
|
0.47 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1
|
|
chr11_-_2162468
Show fit
|
0.45 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr6_-_105585022
Show fit
|
0.45 |
ENST00000314641.5
|
BVES
|
blood vessel epicardial substance
|
|
chr12_-_50616382
Show fit
|
0.39 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr11_-_2162162
Show fit
|
0.39 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr7_-_94285402
Show fit
|
0.38 |
ENST00000428696.2
ENST00000445866.2
|
SGCE
|
sarcoglycan, epsilon
|
|
chr16_-_11375179
Show fit
|
0.38 |
ENST00000312511.3
|
PRM1
|
protamine 1
|
|
chr12_-_50616122
Show fit
|
0.37 |
ENST00000552823.1
ENST00000552909.1
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr7_-_44180884
Show fit
|
0.36 |
ENST00000458240.1
ENST00000223364.3
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr11_-_102323489
Show fit
|
0.35 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123
|
|
chr17_+_73452695
Show fit
|
0.35 |
ENST00000582186.1
ENST00000582455.1
ENST00000581252.1
ENST00000579208.1
|
KIAA0195
|
KIAA0195
|
|
chr3_-_9834375
Show fit
|
0.33 |
ENST00000343450.2
ENST00000301964.2
|
TADA3
|
transcriptional adaptor 3
|
|
chr1_+_11994715
Show fit
|
0.33 |
ENST00000449038.1
ENST00000376369.3
ENST00000429000.2
ENST00000196061.4
|
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr5_-_179050660
Show fit
|
0.30 |
ENST00000519056.1
ENST00000506721.1
ENST00000503105.1
ENST00000504348.1
ENST00000508103.1
ENST00000510431.1
ENST00000515158.1
ENST00000393432.4
ENST00000442819.2
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr7_-_94285472
Show fit
|
0.27 |
ENST00000437425.2
ENST00000447873.1
ENST00000415788.2
|
SGCE
|
sarcoglycan, epsilon
|
|
chr11_-_102323740
Show fit
|
0.27 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123
|
|
chr7_-_94285511
Show fit
|
0.25 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon
|
|
chr18_+_54318566
Show fit
|
0.25 |
ENST00000589935.1
ENST00000357574.3
|
WDR7
|
WD repeat domain 7
|
|
chr17_-_10560619
Show fit
|
0.25 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic
|
|
chr2_-_161056762
Show fit
|
0.24 |
ENST00000428609.2
ENST00000409967.2
|
ITGB6
|
integrin, beta 6
|
|
chr17_-_55911970
Show fit
|
0.23 |
ENST00000581805.1
ENST00000580960.1
|
RP11-60A24.3
|
RP11-60A24.3
|
|
chr10_-_16859361
Show fit
|
0.23 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1
|
|
chr10_-_16859442
Show fit
|
0.23 |
ENST00000602389.1
ENST00000345264.5
|
RSU1
|
Ras suppressor protein 1
|
|
chr16_+_86229728
Show fit
|
0.21 |
ENST00000601250.1
|
LINC01082
|
long intergenic non-protein coding RNA 1082
|
|
chr17_-_4890919
Show fit
|
0.20 |
ENST00000572543.1
ENST00000381311.5
ENST00000348066.3
ENST00000358183.4
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr19_-_15344243
Show fit
|
0.20 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3
|
|
chr1_-_86043921
Show fit
|
0.19 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_151561506
Show fit
|
0.19 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_+_40679584
Show fit
|
0.18 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr19_-_15343773
Show fit
|
0.17 |
ENST00000435261.1
ENST00000594042.1
|
EPHX3
|
epoxide hydrolase 3
|
|
chr17_+_33474860
Show fit
|
0.17 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr19_+_57901208
Show fit
|
0.17 |
ENST00000366197.5
ENST00000596282.1
ENST00000597400.1
ENST00000598895.1
ENST00000336128.7
ENST00000596617.1
|
ZNF548
AC003002.6
|
zinc finger protein 548
Uncharacterized protein
|
|
chr10_+_88428370
Show fit
|
0.15 |
ENST00000372066.3
ENST00000263066.6
ENST00000372056.4
ENST00000310944.6
ENST00000361373.4
ENST00000542786.1
|
LDB3
|
LIM domain binding 3
|
|
chr19_-_35166604
Show fit
|
0.15 |
ENST00000601241.1
|
SCGB2B2
|
secretoglobin, family 2B, member 2
|
|
chrX_-_153602991
Show fit
|
0.14 |
ENST00000369850.3
ENST00000422373.1
|
FLNA
|
filamin A, alpha
|
|
chr5_+_92919043
Show fit
|
0.14 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr17_+_33474826
Show fit
|
0.13 |
ENST00000268876.5
ENST00000433649.1
ENST00000378449.1
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr1_-_168106536
Show fit
|
0.13 |
ENST00000537209.1
ENST00000361697.2
ENST00000546300.1
ENST00000367835.1
|
GPR161
|
G protein-coupled receptor 161
|
|
chr2_+_74120094
Show fit
|
0.12 |
ENST00000409731.3
ENST00000345517.3
ENST00000409918.1
ENST00000442912.1
ENST00000409624.1
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr3_+_187930719
Show fit
|
0.12 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_-_4890649
Show fit
|
0.12 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr2_-_96811170
Show fit
|
0.11 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr8_+_53850991
Show fit
|
0.11 |
ENST00000331251.3
|
NPBWR1
|
neuropeptides B/W receptor 1
|
|
chr15_+_33603147
Show fit
|
0.10 |
ENST00000415757.3
ENST00000389232.4
|
RYR3
|
ryanodine receptor 3
|
|
chr17_-_72358001
Show fit
|
0.09 |
ENST00000375366.3
|
BTBD17
|
BTB (POZ) domain containing 17
|
|
chr17_-_79479789
Show fit
|
0.08 |
ENST00000571691.1
ENST00000571721.1
ENST00000573283.1
ENST00000575842.1
ENST00000575087.1
ENST00000570382.1
ENST00000331925.2
|
ACTG1
|
actin, gamma 1
|
|
chr1_+_223889310
Show fit
|
0.08 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_+_86667117
Show fit
|
0.08 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6
|
|
chr17_-_79481666
Show fit
|
0.08 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1
|
|
chr13_+_27825706
Show fit
|
0.07 |
ENST00000272274.4
ENST00000319826.4
ENST00000326092.4
|
RPL21
|
ribosomal protein L21
|
|
chr11_+_17281900
Show fit
|
0.07 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2
|
|
chr12_-_125398850
Show fit
|
0.06 |
ENST00000535859.1
ENST00000546271.1
ENST00000540700.1
ENST00000546120.1
|
UBC
|
ubiquitin C
|
|
chr12_+_59989918
Show fit
|
0.06 |
ENST00000547379.1
ENST00000549465.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7
|
|
chr7_-_121944491
Show fit
|
0.06 |
ENST00000331178.4
ENST00000427185.2
ENST00000442488.2
|
FEZF1
|
FEZ family zinc finger 1
|
|
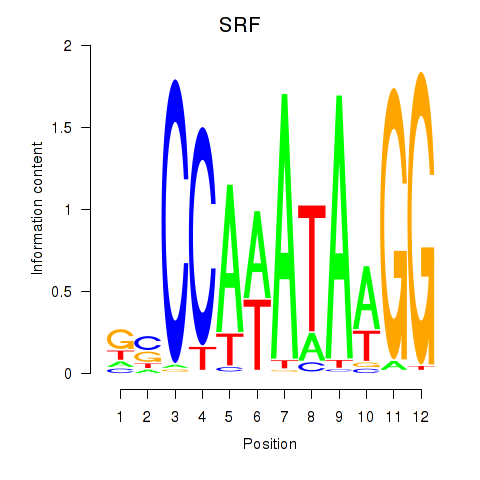
chr6_+_43139037
Show fit
|
0.06 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr3_+_9834227
Show fit
|
0.05 |
ENST00000287613.7
ENST00000397261.3
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr5_+_66254698
Show fit
|
0.04 |
ENST00000405643.1
ENST00000407621.1
ENST00000432426.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr1_+_219347203
Show fit
|
0.03 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1
|
|
chrX_+_22056165
Show fit
|
0.03 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr1_+_219347186
Show fit
|
0.03 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1
|
|
chr16_-_3350614
Show fit
|
0.03 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7
|
|
chr6_-_159466136
Show fit
|
0.03 |
ENST00000367066.3
ENST00000326965.6
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr13_+_27825446
Show fit
|
0.03 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21
|
|
chr17_-_46671323
Show fit
|
0.02 |
ENST00000239151.5
|
HOXB5
|
homeobox B5
|
|
chr4_-_109087872
Show fit
|
0.01 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr18_-_54318353
Show fit
|
0.01 |
ENST00000590954.1
ENST00000540155.1
|
TXNL1
|
thioredoxin-like 1
|
|
chr11_-_65667884
Show fit
|
0.01 |
ENST00000448083.2
ENST00000531493.1
ENST00000532401.1
|
FOSL1
|
FOS-like antigen 1
|
|
chr10_-_17659234
Show fit
|
0.01 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|