Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
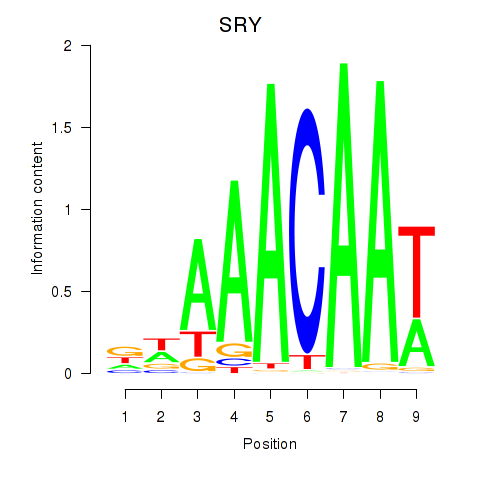
Results for SRY
Z-value: 0.67
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SRY | hg19_v2_chrY_-_2655644_2655740 | -0.25 | 2.3e-01 | Click! |
Activity profile of SRY motif
Sorted Z-values of SRY motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41614909 | 1.90 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41614720 | 1.80 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_-_111091948 | 1.77 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr2_+_228678550 | 1.59 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr14_-_92414055 | 1.47 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr14_-_92413353 | 1.33 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr1_+_61547405 | 1.30 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr13_-_114843416 | 1.10 |
ENST00000389544.4
|
RASA3
|
RAS p21 protein activator 3 |
| chr15_-_59665062 | 1.09 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr11_-_10830463 | 1.02 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_84630053 | 0.97 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_84630645 | 0.94 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_134551583 | 0.89 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr1_-_85930246 | 0.89 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr2_-_208030647 | 0.87 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_92413727 | 0.79 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr1_+_164528866 | 0.77 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr18_+_3448455 | 0.69 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_+_208423891 | 0.68 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr21_+_33784670 | 0.67 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr9_-_14314566 | 0.66 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr9_-_14314518 | 0.66 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr17_-_67138015 | 0.66 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr14_-_51027838 | 0.66 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_57045228 | 0.62 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr18_-_53303123 | 0.59 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chrX_+_84499081 | 0.57 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr15_-_34610962 | 0.57 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr3_-_141868293 | 0.57 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_159409512 | 0.54 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr18_-_52989217 | 0.53 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr2_+_162087577 | 0.53 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_+_53019993 | 0.53 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr14_-_73493784 | 0.52 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr2_-_157198860 | 0.51 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_10829851 | 0.50 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr9_-_123476719 | 0.49 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr1_+_97188188 | 0.48 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr15_-_70390213 | 0.48 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr9_-_123476612 | 0.48 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr14_+_22748980 | 0.47 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr15_-_70994612 | 0.47 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr21_+_17792672 | 0.45 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_-_73493825 | 0.44 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr11_+_128563948 | 0.44 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_140743859 | 0.44 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr14_-_23288930 | 0.43 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr9_-_16727978 | 0.43 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr2_-_192711968 | 0.42 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr2_+_109204743 | 0.42 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_13906250 | 0.41 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr3_-_71353892 | 0.40 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr1_-_160231451 | 0.40 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr8_+_67405755 | 0.39 |
ENST00000521495.1
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr3_+_57882024 | 0.39 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr11_+_128563652 | 0.37 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_15525426 | 0.37 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_+_66797687 | 0.36 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_+_84630367 | 0.36 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_+_13134772 | 0.36 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr18_+_3466248 | 0.35 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr2_-_160473114 | 0.34 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_-_141868357 | 0.33 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr18_-_52969844 | 0.33 |
ENST00000561831.3
|
TCF4
|
transcription factor 4 |
| chr14_+_53019822 | 0.33 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr18_-_53257027 | 0.33 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr14_-_89883412 | 0.32 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chrX_+_84499038 | 0.31 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr18_+_3451584 | 0.31 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_138677515 | 0.30 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr18_+_11851383 | 0.30 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr11_-_111637083 | 0.30 |
ENST00000427203.2
ENST00000341980.6 ENST00000311129.5 ENST00000393055.2 ENST00000426998.2 ENST00000527614.1 |
PPP2R1B
|
protein phosphatase 2, regulatory subunit A, beta |
| chr4_-_76598544 | 0.29 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr20_-_62582475 | 0.29 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr7_+_128399002 | 0.28 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr4_+_170581213 | 0.28 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr3_+_178866199 | 0.27 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr9_+_27109392 | 0.27 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chrX_+_28605516 | 0.27 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_+_140739537 | 0.27 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr2_-_160472952 | 0.27 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_+_205682497 | 0.27 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr22_+_19710468 | 0.26 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr14_+_91580777 | 0.26 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr14_+_91581011 | 0.26 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr2_+_191273052 | 0.26 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr17_-_40428359 | 0.26 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr11_+_128562372 | 0.26 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr13_-_74708372 | 0.24 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr14_+_91580708 | 0.24 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr19_+_13135731 | 0.24 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr3_-_28390298 | 0.24 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr20_-_43150601 | 0.23 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr7_+_29519486 | 0.23 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr8_+_67405794 | 0.23 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr21_-_39870339 | 0.23 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr12_-_81763184 | 0.22 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_+_33116765 | 0.22 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chrX_-_39923656 | 0.22 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr17_-_46716647 | 0.22 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr9_-_39239171 | 0.22 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr11_+_67007518 | 0.21 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr14_+_22985251 | 0.21 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr6_+_108882069 | 0.21 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr7_-_19813192 | 0.20 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr2_+_27665289 | 0.20 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr11_-_82708435 | 0.20 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr18_+_3451646 | 0.20 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_-_11589131 | 0.20 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr17_-_39324424 | 0.20 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr2_-_231989808 | 0.20 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chrX_+_99839799 | 0.19 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr9_-_37465396 | 0.19 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr10_+_111985713 | 0.18 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr5_+_138678131 | 0.17 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr7_+_30185406 | 0.17 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_28390415 | 0.17 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr6_+_84563295 | 0.17 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr12_-_31477072 | 0.17 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr1_-_151119087 | 0.17 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr20_-_3762087 | 0.17 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr11_-_118095801 | 0.17 |
ENST00000356289.5
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr20_-_61002584 | 0.16 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr18_-_52989525 | 0.16 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr14_+_91580732 | 0.16 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr7_-_99698338 | 0.16 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr15_-_52263937 | 0.16 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr9_+_139874683 | 0.15 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr16_-_15736881 | 0.15 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr15_-_86338134 | 0.15 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr14_-_53019211 | 0.15 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr8_+_70404996 | 0.15 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr15_-_86338100 | 0.15 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr4_-_103746683 | 0.14 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_93645818 | 0.14 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr10_+_115312766 | 0.14 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr12_+_60058458 | 0.14 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_+_142985308 | 0.14 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr12_-_56352368 | 0.14 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr11_-_118095718 | 0.14 |
ENST00000526620.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr12_+_130646999 | 0.14 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr2_+_10262857 | 0.14 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr4_-_103746924 | 0.13 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_91451758 | 0.13 |
ENST00000266719.3
|
KERA
|
keratocan |
| chr7_+_29519662 | 0.13 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr17_+_79953310 | 0.13 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr6_+_126070726 | 0.13 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr12_+_50478977 | 0.13 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr12_-_10251539 | 0.12 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr15_-_55562582 | 0.12 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_46272462 | 0.12 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr1_+_185703513 | 0.12 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr6_-_9939552 | 0.12 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr4_-_76598296 | 0.12 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_+_57875711 | 0.11 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr6_-_52109335 | 0.11 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr20_+_10415931 | 0.11 |
ENST00000334534.5
|
SLX4IP
|
SLX4 interacting protein |
| chr5_-_146461027 | 0.11 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr3_-_11610255 | 0.11 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr3_+_28390637 | 0.10 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr4_+_71263599 | 0.10 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr18_+_18943554 | 0.10 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr3_-_28390120 | 0.10 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr5_-_147162078 | 0.10 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr4_-_65275162 | 0.10 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr2_-_37899323 | 0.09 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_+_105404899 | 0.09 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr1_-_217311090 | 0.09 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr12_-_498620 | 0.09 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr13_+_102142296 | 0.09 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_-_178865747 | 0.09 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr13_-_41240717 | 0.09 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr15_-_55562479 | 0.09 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_-_103747011 | 0.08 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_31082603 | 0.08 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr15_-_55563072 | 0.08 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_217250231 | 0.08 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr22_-_36236265 | 0.08 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_28390581 | 0.08 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr9_+_27109133 | 0.08 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr10_+_74653330 | 0.08 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr12_-_102591604 | 0.08 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr10_+_111967345 | 0.08 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr1_+_33722080 | 0.08 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr14_+_91580357 | 0.07 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr2_+_27665232 | 0.07 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr1_+_164529004 | 0.07 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr6_-_32557610 | 0.07 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr16_-_4852915 | 0.07 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr13_-_39564993 | 0.07 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr10_+_11207438 | 0.07 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr1_+_147374915 | 0.07 |
ENST00000240986.4
|
GJA8
|
gap junction protein, alpha 8, 50kDa |
| chr1_+_43855560 | 0.07 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr6_+_139094657 | 0.07 |
ENST00000332797.6
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr8_+_110346546 | 0.06 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr3_+_69812701 | 0.06 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_+_142880512 | 0.06 |
ENST00000446620.1
|
TAS2R39
|
taste receptor, type 2, member 39 |
| chr6_-_87804815 | 0.06 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr6_-_31651817 | 0.06 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr7_-_92855762 | 0.06 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 2.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 3.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 1.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.6 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.7 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.9 | GO:0006527 | arginine catabolic process(GO:0006527) negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 2.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 1.0 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.5 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 2.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 1.0 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 1.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 2.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.7 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |