Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for STAT1_STAT3_BCL6
Z-value: 0.54
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
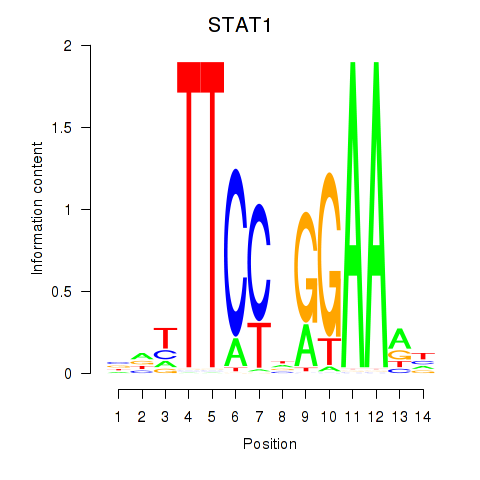
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
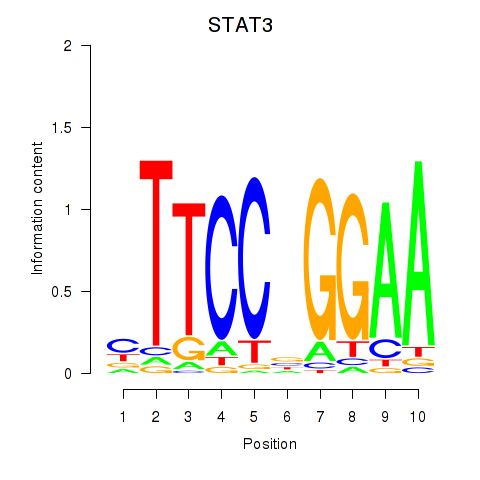
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
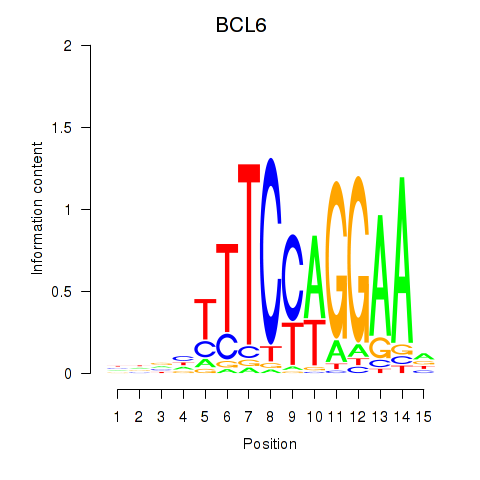
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL6 | hg19_v2_chr3_-_187454281_187454357 | 0.63 | 7.4e-04 | Click! |
| STAT3 | hg19_v2_chr17_-_40540586_40540600 | 0.29 | 1.6e-01 | Click! |
| STAT1 | hg19_v2_chr2_-_191878162_191878313 | -0.05 | 8.0e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_10381769 | 1.80 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr12_+_122064673 | 1.53 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr16_-_67517716 | 1.27 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr17_+_32582293 | 1.11 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr3_-_187455680 | 0.92 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr1_-_41328018 | 0.89 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr5_+_131409476 | 0.81 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr21_+_26934165 | 0.79 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr12_+_93964746 | 0.76 |
ENST00000536696.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_+_53443963 | 0.67 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_+_97203082 | 0.61 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_-_4831701 | 0.61 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr12_+_53443680 | 0.58 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_-_173020056 | 0.49 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr22_-_39636914 | 0.47 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr22_-_39637135 | 0.47 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr1_+_221051699 | 0.44 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr12_-_120687948 | 0.42 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr12_+_122064398 | 0.39 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr3_+_101568349 | 0.39 |
ENST00000326151.5
ENST00000326172.5 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr7_+_2598628 | 0.39 |
ENST00000402050.2
ENST00000404984.1 ENST00000415271.2 ENST00000438376.2 |
IQCE
|
IQ motif containing E |
| chr1_-_153521597 | 0.38 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_+_32683456 | 0.37 |
ENST00000225844.2
|
CCL13
|
chemokine (C-C motif) ligand 13 |
| chr22_-_31503490 | 0.36 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chrX_-_139866723 | 0.36 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr19_-_4540486 | 0.35 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr17_-_26697304 | 0.35 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr4_-_76957214 | 0.34 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr8_-_95274536 | 0.33 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr12_-_9913489 | 0.32 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr19_+_35773242 | 0.32 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr15_+_84322827 | 0.31 |
ENST00000286744.5
ENST00000567476.1 |
ADAMTSL3
|
ADAMTS-like 3 |
| chr15_-_42448788 | 0.31 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr19_-_40324767 | 0.31 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr14_-_81408031 | 0.29 |
ENST00000216517.6
|
CEP128
|
centrosomal protein 128kDa |
| chr18_+_6729698 | 0.28 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr20_-_49308048 | 0.28 |
ENST00000327979.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr1_+_155099927 | 0.26 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr1_-_153521714 | 0.26 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr19_-_40324255 | 0.26 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr22_+_38609538 | 0.25 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr7_-_140178775 | 0.25 |
ENST00000474576.1
ENST00000473444.1 ENST00000471104.1 |
MKRN1
|
makorin ring finger protein 1 |
| chr3_+_187871060 | 0.24 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr17_-_34207295 | 0.23 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr22_+_38597889 | 0.23 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_-_32784687 | 0.22 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr19_+_44081344 | 0.22 |
ENST00000599207.1
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr9_-_134151915 | 0.22 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr19_+_11200038 | 0.22 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr19_+_47835404 | 0.21 |
ENST00000595464.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr6_+_36646435 | 0.21 |
ENST00000244741.5
ENST00000405375.1 ENST00000373711.2 |
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr11_+_46740730 | 0.21 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr17_+_77021702 | 0.20 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr22_+_24990746 | 0.19 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr10_+_104005272 | 0.19 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr5_+_118691706 | 0.19 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_-_64211112 | 0.19 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr17_-_18950310 | 0.19 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr2_+_1488435 | 0.18 |
ENST00000446278.1
ENST00000469607.1 |
TPO
|
thyroid peroxidase |
| chr12_-_42877726 | 0.18 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr7_+_117120017 | 0.17 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr9_-_117880477 | 0.17 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chrX_+_144908928 | 0.17 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr11_+_71710973 | 0.17 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chrX_-_65253506 | 0.17 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr7_-_107443652 | 0.17 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr19_+_50145328 | 0.17 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr11_+_71710648 | 0.17 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr3_+_111393659 | 0.17 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_-_155511887 | 0.16 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr21_-_31744557 | 0.16 |
ENST00000399889.2
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr6_+_30131318 | 0.16 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr17_+_6347729 | 0.16 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr14_+_22458631 | 0.16 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr17_+_6347761 | 0.16 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr7_+_2599056 | 0.16 |
ENST00000325979.7
ENST00000423395.1 |
IQCE
|
IQ motif containing E |
| chrX_-_10544942 | 0.16 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_+_101591314 | 0.16 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr17_+_36858694 | 0.15 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr14_-_30396948 | 0.15 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr22_+_31518938 | 0.15 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr4_+_69313145 | 0.15 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr3_-_119379427 | 0.15 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr9_+_125796806 | 0.14 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr12_+_56618102 | 0.14 |
ENST00000267023.4
ENST00000380198.2 ENST00000341463.5 |
NABP2
|
nucleic acid binding protein 2 |
| chr1_-_168464875 | 0.14 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_+_7598890 | 0.14 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr4_-_76823681 | 0.14 |
ENST00000286719.7
|
PPEF2
|
protein phosphatase, EF-hand calcium binding domain 2 |
| chr1_-_11107280 | 0.13 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr4_+_126237554 | 0.13 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr3_+_111393501 | 0.13 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr19_+_55327923 | 0.13 |
ENST00000391728.4
ENST00000326542.7 ENST00000358178.4 |
KIR3DL1
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 |
| chr1_-_203198790 | 0.13 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr20_-_18774614 | 0.13 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr11_+_64059464 | 0.13 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr7_+_74508372 | 0.13 |
ENST00000356115.5
ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr11_+_6226782 | 0.13 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr8_+_145726472 | 0.13 |
ENST00000528430.1
|
PPP1R16A
|
protein phosphatase 1, regulatory subunit 16A |
| chr12_+_66696322 | 0.12 |
ENST00000247815.4
|
HELB
|
helicase (DNA) B |
| chr2_+_202122703 | 0.12 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr6_-_33548006 | 0.12 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr16_-_67260691 | 0.12 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr16_-_74734672 | 0.12 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr6_-_33547975 | 0.12 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr2_-_105030466 | 0.12 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr14_+_24779376 | 0.12 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr15_-_72523924 | 0.12 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr12_-_88974236 | 0.12 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr15_+_74165945 | 0.12 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr17_-_76274572 | 0.12 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr22_+_20748456 | 0.11 |
ENST00000420626.1
ENST00000356671.5 |
ZNF74
|
zinc finger protein 74 |
| chr19_+_13229126 | 0.11 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr22_-_50523760 | 0.11 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_-_126327398 | 0.11 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr10_+_6625733 | 0.11 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr19_+_18530146 | 0.11 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr19_+_49535169 | 0.11 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr12_-_48213568 | 0.11 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr4_-_140477353 | 0.11 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_-_75443118 | 0.11 |
ENST00000222902.2
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr8_+_22462145 | 0.11 |
ENST00000308511.4
ENST00000523801.1 ENST00000521301.1 |
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr4_-_2758015 | 0.10 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr12_-_7261772 | 0.10 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr9_-_123239632 | 0.10 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr14_-_25479811 | 0.10 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr1_-_154150651 | 0.10 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr3_-_123512688 | 0.10 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_61197572 | 0.10 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr14_+_23842018 | 0.10 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr14_-_81408093 | 0.10 |
ENST00000555265.1
|
CEP128
|
centrosomal protein 128kDa |
| chr11_-_19223523 | 0.10 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_-_111782484 | 0.10 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr8_-_82395461 | 0.09 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr19_+_7599128 | 0.09 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr2_-_79386786 | 0.09 |
ENST00000393878.1
ENST00000305165.2 ENST00000409839.3 |
REG3A
|
regenerating islet-derived 3 alpha |
| chr3_+_44840679 | 0.09 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr14_+_94577074 | 0.09 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr6_+_116782527 | 0.09 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr19_+_7599597 | 0.09 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr19_-_6737241 | 0.09 |
ENST00000430424.4
ENST00000597298.1 |
GPR108
|
G protein-coupled receptor 108 |
| chr18_-_5396271 | 0.09 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr18_+_6729725 | 0.09 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr18_+_23806437 | 0.09 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr15_+_75182346 | 0.09 |
ENST00000569931.1
ENST00000352410.4 ENST00000566377.1 ENST00000569233.1 ENST00000567132.1 ENST00000564633.1 ENST00000568907.1 ENST00000563422.1 ENST00000564003.1 ENST00000562800.1 ENST00000563786.1 ENST00000535694.1 ENST00000323744.6 ENST00000568828.1 ENST00000562606.1 ENST00000565576.1 ENST00000567570.1 |
MPI
|
mannose phosphate isomerase |
| chr20_+_23331373 | 0.09 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr6_-_117150198 | 0.09 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr22_+_33197683 | 0.09 |
ENST00000266085.6
|
TIMP3
|
TIMP metallopeptidase inhibitor 3 |
| chr16_-_46782221 | 0.09 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr19_-_6737576 | 0.09 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chr12_+_13197218 | 0.09 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr11_+_57365150 | 0.09 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr1_+_9294822 | 0.09 |
ENST00000377403.2
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr4_+_158141899 | 0.08 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr11_-_61197480 | 0.08 |
ENST00000439958.3
ENST00000394888.4 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chrX_-_70473957 | 0.08 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr18_-_10748498 | 0.08 |
ENST00000579949.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr1_-_150780757 | 0.08 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr13_+_111767650 | 0.08 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_+_153004800 | 0.08 |
ENST00000392661.3
|
SPRR1B
|
small proline-rich protein 1B |
| chr12_-_56352368 | 0.08 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr17_+_32646055 | 0.08 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chr14_-_25519095 | 0.08 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr10_-_25010795 | 0.08 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr17_-_73851285 | 0.08 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr4_+_155484155 | 0.08 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_+_142031986 | 0.08 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr1_-_154127518 | 0.08 |
ENST00000368559.3
ENST00000271854.3 |
NUP210L
|
nucleoporin 210kDa-like |
| chr6_-_43484621 | 0.08 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr19_-_44304167 | 0.08 |
ENST00000594013.1
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr16_-_3030407 | 0.08 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr5_+_140579162 | 0.08 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr6_+_32821924 | 0.08 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_-_42877764 | 0.08 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr11_+_60163775 | 0.08 |
ENST00000300187.6
ENST00000395005.2 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr2_+_37423618 | 0.07 |
ENST00000402297.1
ENST00000397064.2 ENST00000406711.1 ENST00000392061.2 ENST00000397226.2 |
AC007390.5
|
CEBPZ antisense RNA 1 |
| chr2_-_84686552 | 0.07 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr18_-_44181442 | 0.07 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr1_-_200379180 | 0.07 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr4_-_21546272 | 0.07 |
ENST00000509207.1
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr3_-_42744312 | 0.07 |
ENST00000416756.1
ENST00000441594.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr1_+_17906970 | 0.07 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr1_-_155006224 | 0.07 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr19_+_2476116 | 0.07 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr7_-_100881041 | 0.07 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr3_+_121613265 | 0.07 |
ENST00000295605.2
|
SLC15A2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr11_+_5646213 | 0.07 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr17_+_40912764 | 0.07 |
ENST00000589683.1
ENST00000588928.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr11_+_60163918 | 0.07 |
ENST00000526375.1
ENST00000531783.1 ENST00000395001.1 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr19_+_42746927 | 0.07 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr1_+_248185250 | 0.07 |
ENST00000355281.1
|
OR2L5
|
olfactory receptor, family 2, subfamily L, member 5 |
| chr3_+_183894566 | 0.07 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr14_-_107083690 | 0.07 |
ENST00000455737.1
ENST00000390629.2 |
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr2_-_99917639 | 0.07 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr20_-_43280361 | 0.07 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr6_+_159290917 | 0.07 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr2_-_175499294 | 0.07 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr16_-_75498308 | 0.07 |
ENST00000569540.1
|
TMEM170A
|
transmembrane protein 170A |
| chr20_-_25320367 | 0.07 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr1_+_171810606 | 0.07 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr19_+_49128209 | 0.07 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 1.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.3 | 0.9 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 0.9 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.2 | 0.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.5 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 1.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.3 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.6 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.2 | GO:2000110 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.3 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 0.2 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.2 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.0 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.0 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0030222 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.4 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.3 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 2.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.9 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |