Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
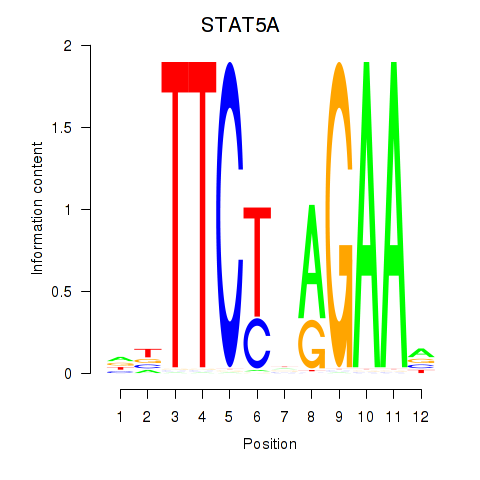
Results for STAT5A
Z-value: 1.04
Transcription factors associated with STAT5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5A
|
ENSG00000126561.12 | signal transducer and activator of transcription 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5A | hg19_v2_chr17_+_40440481_40440561 | 0.71 | 7.0e-05 | Click! |
Activity profile of STAT5A motif
Sorted Z-values of STAT5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_152214098 | 3.78 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr11_-_102668879 | 3.57 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr10_-_49812997 | 3.19 |
ENST00000417912.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_+_10381769 | 3.04 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr10_-_6019552 | 2.85 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr15_+_67458357 | 2.84 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr6_-_32820529 | 2.69 |
ENST00000425148.2
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_-_153521714 | 2.66 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr4_+_142558078 | 2.65 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr4_+_142557771 | 2.61 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr3_+_122399444 | 2.56 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr2_+_102972363 | 2.54 |
ENST00000409599.1
|
IL18R1
|
interleukin 18 receptor 1 |
| chr15_+_67430339 | 2.38 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr13_-_43566301 | 2.33 |
ENST00000398762.3
ENST00000313640.7 ENST00000313624.7 |
EPSTI1
|
epithelial stromal interaction 1 (breast) |
| chr1_-_153521597 | 2.32 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr10_-_49813090 | 2.29 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr1_-_8000872 | 2.28 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr4_-_76957214 | 2.25 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr1_-_41328018 | 2.06 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr21_-_45660723 | 2.03 |
ENST00000344330.4
ENST00000407780.3 ENST00000400379.3 |
ICOSLG
|
inducible T-cell co-stimulator ligand |
| chr12_+_102271129 | 1.99 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_-_173020056 | 1.91 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr8_+_86157699 | 1.80 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr17_+_77021702 | 1.73 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr21_+_42792442 | 1.72 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr4_-_120550146 | 1.69 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr11_+_69455855 | 1.64 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr17_+_41158742 | 1.63 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr12_+_21207503 | 1.63 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr7_+_55177416 | 1.59 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr1_-_89488510 | 1.56 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr17_+_40440481 | 1.50 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr19_+_18284477 | 1.49 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr21_+_26934165 | 1.44 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr19_-_55672037 | 1.41 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr6_+_4706368 | 1.36 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr10_-_6019455 | 1.35 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr3_-_79816965 | 1.31 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr16_-_75590114 | 1.30 |
ENST00000568377.1
ENST00000565067.1 ENST00000258173.6 |
TMEM231
|
transmembrane protein 231 |
| chr17_-_33864772 | 1.26 |
ENST00000361112.4
|
SLFN12L
|
schlafen family member 12-like |
| chr19_+_57106647 | 1.26 |
ENST00000328070.6
|
ZNF71
|
zinc finger protein 71 |
| chr3_-_123512688 | 1.24 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr1_+_17906970 | 1.24 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr22_+_31644388 | 1.24 |
ENST00000333611.4
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr8_+_23386305 | 1.22 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr5_-_13944652 | 1.22 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr22_+_31608219 | 1.20 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr19_+_10197463 | 1.20 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_32853093 | 1.18 |
ENST00000448773.1
ENST00000317907.4 |
TTC27
|
tetratricopeptide repeat domain 27 |
| chr5_+_127419449 | 1.16 |
ENST00000262461.2
ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr18_+_55816546 | 1.13 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_-_105892937 | 1.12 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr10_+_115438920 | 1.11 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr16_-_11681316 | 1.09 |
ENST00000571688.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr1_-_144994840 | 1.08 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr14_+_22538811 | 1.06 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr7_+_6144514 | 1.06 |
ENST00000306177.5
ENST00000465073.2 |
USP42
|
ubiquitin specific peptidase 42 |
| chr16_+_50730910 | 1.05 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr12_+_133657461 | 1.03 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr7_-_108096822 | 1.02 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr10_+_115439282 | 1.02 |
ENST00000369321.2
ENST00000345633.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr19_+_42724423 | 1.01 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr16_-_75498308 | 1.01 |
ENST00000569540.1
|
TMEM170A
|
transmembrane protein 170A |
| chr17_-_34207295 | 1.00 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr1_-_55230165 | 1.00 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr10_+_115439630 | 0.99 |
ENST00000369318.3
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr15_+_63414017 | 0.97 |
ENST00000413507.2
|
LACTB
|
lactamase, beta |
| chr21_+_34602680 | 0.97 |
ENST00000447980.1
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr16_+_57653854 | 0.96 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_+_115439699 | 0.95 |
ENST00000369315.1
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr10_+_6625605 | 0.95 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr17_+_77030267 | 0.94 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_-_117318788 | 0.94 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr6_+_116692102 | 0.91 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr15_+_63414760 | 0.90 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr8_+_38586068 | 0.90 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr4_-_185395672 | 0.90 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr6_-_137539651 | 0.88 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr6_+_108487245 | 0.88 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr18_-_44561988 | 0.88 |
ENST00000332567.4
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2) |
| chr1_-_145075847 | 0.87 |
ENST00000530740.1
ENST00000369359.4 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_-_30654977 | 0.86 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr8_-_12612962 | 0.86 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chrX_+_1455478 | 0.86 |
ENST00000331035.4
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr7_-_108096765 | 0.84 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr5_+_149887672 | 0.83 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr9_+_98534605 | 0.81 |
ENST00000600140.1
|
DKFZP434H0512
|
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
| chr22_-_50523760 | 0.81 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_+_20968608 | 0.81 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_+_201994569 | 0.78 |
ENST00000457277.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr14_-_91710852 | 0.76 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chrX_+_149861836 | 0.76 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr7_+_7606497 | 0.76 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr15_-_70387120 | 0.76 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr10_+_90750378 | 0.76 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chrX_-_11445856 | 0.75 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr18_+_61637159 | 0.74 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr16_+_3313791 | 0.72 |
ENST00000538765.1
|
ZNF263
|
zinc finger protein 263 |
| chr1_-_201438282 | 0.72 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr14_-_51135036 | 0.72 |
ENST00000324679.4
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr22_-_32651326 | 0.71 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr22_-_31503490 | 0.71 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr19_+_44081344 | 0.69 |
ENST00000599207.1
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr18_-_74839891 | 0.68 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr12_+_10658489 | 0.68 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr20_+_58251716 | 0.68 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr8_+_27629459 | 0.66 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr15_-_38856836 | 0.65 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr12_-_10151773 | 0.65 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr21_+_34602200 | 0.65 |
ENST00000382264.3
ENST00000382241.3 ENST00000404220.3 ENST00000342136.4 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr21_+_34602377 | 0.64 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr5_+_121297650 | 0.64 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr7_+_117120017 | 0.63 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr1_-_144994909 | 0.63 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_+_72058130 | 0.63 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_-_11807277 | 0.63 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_+_35168547 | 0.63 |
ENST00000502743.1
ENST00000509528.1 ENST00000506901.1 |
ZNF302
|
zinc finger protein 302 |
| chr2_-_208634287 | 0.62 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr14_-_24898731 | 0.62 |
ENST00000267406.6
|
CBLN3
|
cerebellin 3 precursor |
| chr4_+_169418195 | 0.60 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_10658201 | 0.60 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr22_+_21128167 | 0.59 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr1_+_144146808 | 0.59 |
ENST00000369190.5
ENST00000412624.2 ENST00000369365.3 |
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr6_-_159240415 | 0.59 |
ENST00000367075.3
|
EZR
|
ezrin |
| chr9_+_103189660 | 0.59 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr11_-_104827425 | 0.57 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_85636928 | 0.57 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_-_28113217 | 0.57 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr22_-_18507279 | 0.57 |
ENST00000441493.2
ENST00000444520.1 ENST00000207726.7 ENST00000429452.1 |
MICAL3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr6_+_108977520 | 0.57 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr5_-_59783882 | 0.56 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr15_-_72767490 | 0.56 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr1_-_160924589 | 0.56 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr6_+_144471643 | 0.56 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr10_+_90750493 | 0.55 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr10_+_114043493 | 0.55 |
ENST00000369422.3
|
TECTB
|
tectorin beta |
| chr21_-_37451680 | 0.54 |
ENST00000399201.1
|
SETD4
|
SET domain containing 4 |
| chr7_+_112063192 | 0.53 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr1_+_168756151 | 0.53 |
ENST00000420691.1
|
LINC00626
|
long intergenic non-protein coding RNA 626 |
| chr2_+_108994466 | 0.53 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr22_+_37257015 | 0.53 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr20_-_43280361 | 0.53 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr3_+_149191723 | 0.52 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr11_-_133715394 | 0.52 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr22_-_45636650 | 0.52 |
ENST00000336156.5
|
KIAA0930
|
KIAA0930 |
| chr9_-_94877658 | 0.52 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr22_-_47882857 | 0.51 |
ENST00000405369.3
|
LL22NC03-75H12.2
|
Novel protein; Uncharacterized protein |
| chr8_-_128960591 | 0.51 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr11_-_59383617 | 0.51 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr12_+_22199108 | 0.51 |
ENST00000229329.2
|
CMAS
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr3_+_151531810 | 0.51 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr7_-_26578407 | 0.51 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr12_+_22778291 | 0.50 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr9_+_124922171 | 0.50 |
ENST00000373764.3
ENST00000536616.1 |
MORN5
|
MORN repeat containing 5 |
| chr2_-_152146385 | 0.50 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr19_-_58090240 | 0.50 |
ENST00000196489.3
|
ZNF416
|
zinc finger protein 416 |
| chr11_-_104905840 | 0.50 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr11_+_58912240 | 0.50 |
ENST00000527629.1
ENST00000361723.3 ENST00000531408.1 |
FAM111A
|
family with sequence similarity 111, member A |
| chr12_-_69080590 | 0.49 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chrX_+_122993657 | 0.49 |
ENST00000434753.3
ENST00000430625.1 |
XIAP
|
X-linked inhibitor of apoptosis |
| chr16_+_56672571 | 0.49 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr2_+_108994633 | 0.49 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr10_-_27443294 | 0.48 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr11_+_117070904 | 0.48 |
ENST00000529792.1
|
TAGLN
|
transgelin |
| chr15_+_66797627 | 0.48 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr21_+_34697209 | 0.48 |
ENST00000270139.3
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr19_-_6737241 | 0.48 |
ENST00000430424.4
ENST00000597298.1 |
GPR108
|
G protein-coupled receptor 108 |
| chr1_-_144995002 | 0.48 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_-_4758159 | 0.48 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr3_-_107941209 | 0.47 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr10_-_27443155 | 0.47 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chr1_+_114471809 | 0.47 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chrX_+_18725758 | 0.47 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr6_-_33548006 | 0.46 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr1_-_100643765 | 0.46 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr17_-_15502111 | 0.46 |
ENST00000354433.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr11_-_9025541 | 0.46 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr1_-_153283194 | 0.45 |
ENST00000290722.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr6_-_33547975 | 0.45 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr2_-_69098566 | 0.45 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr9_-_136203235 | 0.45 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr11_-_117695449 | 0.45 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr1_-_23342340 | 0.44 |
ENST00000566855.1
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr15_+_101142722 | 0.44 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr5_+_8457793 | 0.44 |
ENST00000606459.1
|
MIR4458HG
|
MIR4458 host gene (non-protein coding) |
| chr10_+_82298088 | 0.44 |
ENST00000470604.2
|
SH2D4B
|
SH2 domain containing 4B |
| chr20_+_61436146 | 0.44 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr16_-_21663919 | 0.44 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr12_+_70132632 | 0.44 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr19_+_44507091 | 0.44 |
ENST00000429154.2
ENST00000585632.1 |
ZNF230
|
zinc finger protein 230 |
| chr1_-_89357179 | 0.43 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr15_+_66797455 | 0.43 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr1_-_144995074 | 0.43 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr3_+_111260980 | 0.43 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr6_+_7541808 | 0.43 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr7_+_134464376 | 0.43 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr4_+_155484155 | 0.43 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr14_+_22320634 | 0.43 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr11_-_62609281 | 0.43 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr1_+_171454639 | 0.42 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT5A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 1.0 | 5.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.8 | 3.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.7 | 2.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 4.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.5 | 1.9 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.4 | 1.3 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.4 | 2.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.4 | 1.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.4 | 1.6 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.4 | 1.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 1.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 2.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.3 | 2.7 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.3 | 1.0 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.3 | 0.9 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 0.9 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 2.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 1.9 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.2 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 2.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.2 | 0.6 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.6 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 1.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 0.5 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 0.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 0.5 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 1.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.8 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 0.5 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 1.8 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.5 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.9 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.6 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 1.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.4 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 3.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.4 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 1.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.7 | GO:1904207 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.4 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.7 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.3 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 1.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 2.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 3.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:1905026 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 1.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 2.6 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.1 | 0.2 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.2 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.7 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.4 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.3 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.1 | 0.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.3 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 2.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.3 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.5 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0071315 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.0 | 1.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 1.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.4 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 1.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.5 | GO:0044117 | growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.4 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.6 | GO:0002715 | regulation of natural killer cell mediated immunity(GO:0002715) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 1.3 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 1.6 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 2.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 1.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.1 | 0.6 | GO:0097454 | microspike(GO:0044393) Schwann cell microvillus(GO:0097454) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 4.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 2.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0035580 | specific granule lumen(GO:0035580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.5 | 2.6 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.4 | 2.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 2.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.3 | 1.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.3 | 1.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.3 | 1.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 1.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 0.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 0.9 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 2.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 2.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.1 | 4.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 4.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.4 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.2 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.4 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 2.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.4 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 4.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 9.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 4.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 16.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 7.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 3.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 5.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 4.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |