Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
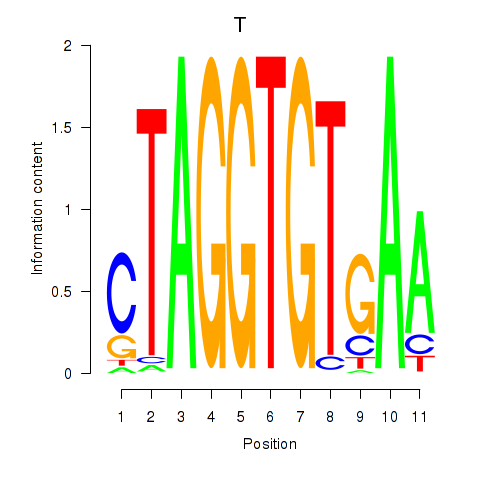
Results for T
Z-value: 0.47
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSG00000164458.5 | T |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | hg19_v2_chr6_-_166582107_166582188 | -0.17 | 4.2e-01 | Click! |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102759199 | 1.30 |
ENST00000409288.1
ENST00000410023.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr1_-_205290865 | 1.13 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr19_-_6690723 | 1.02 |
ENST00000601008.1
|
C3
|
complement component 3 |
| chr19_+_10197463 | 0.93 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr17_+_26662597 | 0.90 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_-_20370847 | 0.83 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr12_+_22778291 | 0.80 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr3_+_127770455 | 0.68 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr19_-_44174330 | 0.67 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_-_74497432 | 0.60 |
ENST00000590288.1
ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr12_-_117318788 | 0.57 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr5_+_14664762 | 0.54 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr11_-_85779971 | 0.49 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_+_54320078 | 0.46 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr2_+_171571827 | 0.46 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr2_-_152146385 | 0.44 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr19_-_44174305 | 0.44 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr2_+_162087577 | 0.42 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_85930246 | 0.42 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr21_+_25801041 | 0.42 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr3_-_66551351 | 0.41 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr2_-_208030647 | 0.39 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_66551397 | 0.39 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr17_+_19281034 | 0.36 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_+_83073334 | 0.36 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr2_+_149894968 | 0.34 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr2_+_149895207 | 0.33 |
ENST00000409876.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr19_-_10514184 | 0.32 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr20_+_32951070 | 0.32 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr1_+_114472481 | 0.31 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr19_+_7030589 | 0.31 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chr7_-_142176790 | 0.30 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr15_+_60296421 | 0.30 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr1_+_114471809 | 0.29 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr19_-_1021113 | 0.29 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr17_-_26662440 | 0.29 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr12_+_54379569 | 0.28 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr19_-_17958832 | 0.27 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr17_-_26662464 | 0.26 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_+_7049332 | 0.26 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
| chr7_+_134576317 | 0.26 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr17_-_19281203 | 0.26 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr17_+_34431212 | 0.26 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chrX_+_16804544 | 0.25 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr3_-_52443799 | 0.24 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr20_+_3190006 | 0.24 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr10_-_97050777 | 0.23 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr1_-_26633067 | 0.23 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chrX_+_153665248 | 0.23 |
ENST00000447750.2
|
GDI1
|
GDP dissociation inhibitor 1 |
| chr7_-_81399411 | 0.22 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_+_95078714 | 0.22 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_-_49351228 | 0.21 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr17_+_17942684 | 0.21 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr7_-_994302 | 0.20 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr1_+_27648648 | 0.20 |
ENST00000374076.4
|
TMEM222
|
transmembrane protein 222 |
| chr3_+_195413160 | 0.20 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr17_+_7533439 | 0.20 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr1_+_27648709 | 0.19 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr12_+_57610562 | 0.18 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr12_-_53074182 | 0.18 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr12_-_49351303 | 0.18 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr19_+_11546440 | 0.18 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr7_-_28220354 | 0.17 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr16_-_2314222 | 0.17 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr19_+_55281260 | 0.17 |
ENST00000336077.6
ENST00000291633.7 |
KIR2DL1
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1 |
| chr17_+_34430980 | 0.17 |
ENST00000250151.4
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr2_+_223162866 | 0.17 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr2_+_108905325 | 0.16 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr1_+_114472222 | 0.16 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr7_+_110731062 | 0.16 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr22_+_35776828 | 0.16 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr20_+_32951041 | 0.16 |
ENST00000374864.4
|
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr19_+_11546153 | 0.16 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr6_+_131958436 | 0.16 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_+_39456895 | 0.16 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr5_-_112257914 | 0.15 |
ENST00000513339.1
ENST00000545426.1 ENST00000504247.1 |
REEP5
|
receptor accessory protein 5 |
| chr2_+_108905095 | 0.15 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_-_38370536 | 0.15 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr5_-_137610300 | 0.14 |
ENST00000274721.3
|
GFRA3
|
GDNF family receptor alpha 3 |
| chr5_-_137610254 | 0.14 |
ENST00000378362.3
|
GFRA3
|
GDNF family receptor alpha 3 |
| chr6_+_29079668 | 0.14 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr17_+_34538310 | 0.14 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr11_-_66112555 | 0.14 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr10_+_4868460 | 0.13 |
ENST00000532248.1
ENST00000345253.5 ENST00000334019.4 |
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr9_-_14722715 | 0.13 |
ENST00000380911.3
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr19_+_55235969 | 0.13 |
ENST00000402254.2
ENST00000538269.1 ENST00000541392.1 ENST00000291860.1 ENST00000396284.2 |
KIR3DL1
KIR3DL3
KIR2DL4
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr7_-_76255444 | 0.13 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_+_92417716 | 0.13 |
ENST00000402388.1
|
BRDT
|
bromodomain, testis-specific |
| chr2_+_233320827 | 0.13 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr6_-_26124138 | 0.12 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr19_+_11546093 | 0.12 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr17_-_74533734 | 0.12 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr1_+_89150245 | 0.11 |
ENST00000370513.5
|
PKN2
|
protein kinase N2 |
| chr1_-_24126051 | 0.11 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr5_+_17444119 | 0.11 |
ENST00000503267.1
ENST00000504998.1 |
RP11-321E2.4
|
RP11-321E2.4 |
| chr3_-_52002403 | 0.11 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr4_+_26322409 | 0.11 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr9_+_36572851 | 0.11 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr9_-_117150303 | 0.11 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr1_+_35225339 | 0.11 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr21_+_19617140 | 0.11 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr5_+_50678921 | 0.11 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr12_-_6961050 | 0.10 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr3_+_133502877 | 0.10 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr7_+_107110488 | 0.10 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr10_-_79789291 | 0.10 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr6_+_24126350 | 0.09 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr20_+_1115821 | 0.09 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_-_24126023 | 0.09 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr1_+_78245303 | 0.09 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr4_+_123300664 | 0.09 |
ENST00000388725.2
|
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr13_-_39564993 | 0.09 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr17_+_34640031 | 0.09 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr4_+_26322185 | 0.08 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_14058794 | 0.08 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr4_+_128554081 | 0.08 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr5_-_131330272 | 0.08 |
ENST00000379240.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_+_166152283 | 0.08 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr12_-_69326940 | 0.07 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr21_-_45078019 | 0.07 |
ENST00000542962.1
|
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr3_+_167453493 | 0.07 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr5_+_140557371 | 0.07 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr1_+_114471972 | 0.07 |
ENST00000369559.4
ENST00000369554.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr10_-_115423792 | 0.07 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr7_-_81399329 | 0.07 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_+_65672702 | 0.06 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr1_+_203765437 | 0.06 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr17_+_34639793 | 0.06 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr7_+_117864815 | 0.06 |
ENST00000433239.1
|
ANKRD7
|
ankyrin repeat domain 7 |
| chr8_+_7801144 | 0.06 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr7_-_87849340 | 0.06 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr20_-_1309809 | 0.06 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_+_122494676 | 0.06 |
ENST00000455432.1
|
TSN
|
translin |
| chr17_+_57970469 | 0.06 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr22_-_41215291 | 0.06 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr18_-_77276057 | 0.05 |
ENST00000597412.1
|
AC018445.1
|
Uncharacterized protein |
| chr15_-_34502197 | 0.05 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr7_-_81399438 | 0.05 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr8_-_141810634 | 0.05 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr15_-_83474806 | 0.05 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr20_+_1093891 | 0.05 |
ENST00000333082.3
ENST00000381898.4 ENST00000381899.4 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr6_+_159290917 | 0.05 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr5_-_131329918 | 0.05 |
ENST00000357096.1
ENST00000431707.1 ENST00000434099.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr11_-_44972476 | 0.05 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr5_+_139554227 | 0.05 |
ENST00000261811.4
|
CYSTM1
|
cysteine-rich transmembrane module containing 1 |
| chr1_-_204135450 | 0.04 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr19_+_3721719 | 0.04 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr4_-_69817481 | 0.04 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr6_-_29013017 | 0.04 |
ENST00000377175.1
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1 |
| chr12_+_128399965 | 0.04 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr19_-_18366182 | 0.04 |
ENST00000355502.3
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr10_+_51565188 | 0.04 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr13_+_24144509 | 0.04 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_133874577 | 0.04 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr1_-_156542328 | 0.04 |
ENST00000361170.2
|
IQGAP3
|
IQ motif containing GTPase activating protein 3 |
| chr7_+_117864708 | 0.03 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr11_-_61197187 | 0.03 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr17_-_56084578 | 0.03 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr4_-_73434498 | 0.03 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr16_+_72459838 | 0.03 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr3_-_133614597 | 0.03 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr14_-_107211459 | 0.03 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chrX_+_79591003 | 0.03 |
ENST00000538312.1
|
FAM46D
|
family with sequence similarity 46, member D |
| chr7_-_229557 | 0.02 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr16_+_69221028 | 0.02 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr7_-_752577 | 0.02 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr22_-_36031181 | 0.02 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr15_-_83621435 | 0.02 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr2_-_85636928 | 0.02 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr11_+_133938955 | 0.02 |
ENST00000534549.1
ENST00000441717.3 |
JAM3
|
junctional adhesion molecule 3 |
| chr3_-_57678772 | 0.02 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr1_+_101702417 | 0.02 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr8_-_114449112 | 0.02 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr20_+_34129770 | 0.02 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr4_-_186456652 | 0.02 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr1_+_33219592 | 0.02 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr15_-_81195510 | 0.02 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr16_-_88717423 | 0.02 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr3_+_102153859 | 0.02 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr3_+_38307293 | 0.01 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr14_-_69619689 | 0.01 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr4_-_186456766 | 0.01 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr12_-_6960407 | 0.01 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr11_+_133938820 | 0.01 |
ENST00000299106.4
ENST00000529443.2 |
JAM3
|
junctional adhesion molecule 3 |
| chr5_+_85913721 | 0.01 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr4_-_140005443 | 0.01 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr4_-_151936865 | 0.00 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_-_56709674 | 0.00 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr6_-_106773291 | 0.00 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr12_+_128399917 | 0.00 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr12_-_52867569 | 0.00 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr1_-_204165610 | 0.00 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 1.0 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.3 | 1.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 0.7 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0035519 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.8 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.2 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.5 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.4 | GO:0051342 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.2 | GO:0032764 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.1 | GO:0071655 | granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.3 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.3 | 1.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |