Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
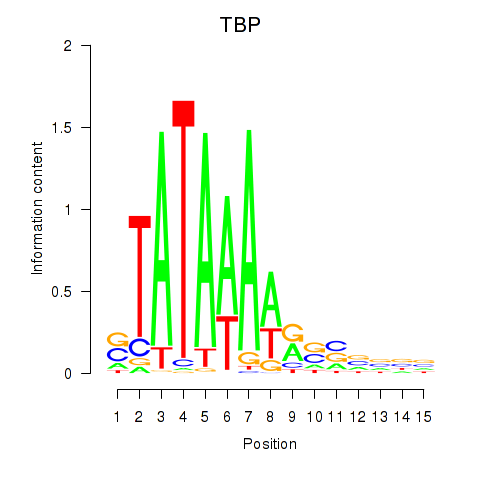
Results for TBP
Z-value: 1.53
Transcription factors associated with TBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBP
|
ENSG00000112592.8 | TATA-box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBP | hg19_v2_chr6_+_170863421_170863484 | -0.29 | 1.5e-01 | Click! |
Activity profile of TBP motif
Sorted Z-values of TBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_169703203 | 15.51 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr19_+_10381769 | 12.19 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr16_+_57406368 | 10.78 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr11_-_102651343 | 8.84 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_+_74606223 | 7.63 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr15_+_62359175 | 5.30 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium-dependent domain containing 4A |
| chr2_+_152214098 | 5.15 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr16_+_56685796 | 5.03 |
ENST00000334346.2
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr4_+_74735102 | 4.82 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr4_+_74702214 | 4.80 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr4_-_74904398 | 4.69 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr14_-_55369525 | 4.35 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr1_-_95007193 | 3.81 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr15_-_62457480 | 3.28 |
ENST00000380392.3
|
C2CD4B
|
C2 calcium-dependent domain containing 4B |
| chr10_+_91087651 | 3.06 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_-_92777606 | 3.05 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr3_+_101546827 | 2.92 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr17_-_34207295 | 2.73 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr1_-_89488510 | 2.71 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr11_+_43964055 | 2.66 |
ENST00000528572.1
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr17_+_77021702 | 2.66 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr18_+_21718924 | 2.55 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr4_+_103422471 | 2.54 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr18_+_21719018 | 2.44 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr19_+_6531010 | 2.37 |
ENST00000245817.3
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9 |
| chr16_+_56703737 | 2.37 |
ENST00000569155.1
|
MT1H
|
metallothionein 1H |
| chr2_-_208031542 | 2.34 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr21_-_35899113 | 2.30 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr6_-_134495992 | 2.30 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_-_24770632 | 2.27 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr16_+_56703703 | 2.26 |
ENST00000332374.4
|
MT1H
|
metallothionein 1H |
| chr8_+_86089460 | 2.24 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr19_+_49375649 | 2.22 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_+_181057638 | 2.21 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr11_-_64646086 | 2.21 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1 |
| chr16_+_56659687 | 2.20 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr2_-_208030647 | 2.15 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_-_3227877 | 2.12 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr13_-_43566301 | 2.08 |
ENST00000398762.3
ENST00000313640.7 ENST00000313624.7 |
EPSTI1
|
epithelial stromal interaction 1 (breast) |
| chr17_+_32612687 | 2.07 |
ENST00000305869.3
|
CCL11
|
chemokine (C-C motif) ligand 11 |
| chr1_+_37940153 | 2.05 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr8_-_23261589 | 2.03 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr8_+_66955648 | 1.98 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr17_+_32646055 | 1.96 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chr14_+_65007177 | 1.96 |
ENST00000247207.6
|
HSPA2
|
heat shock 70kDa protein 2 |
| chr6_-_137539651 | 1.86 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr11_-_2950642 | 1.82 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr5_+_179247759 | 1.77 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr16_-_56701933 | 1.76 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr1_-_209825674 | 1.73 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr5_-_169816638 | 1.73 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr2_-_175711133 | 1.70 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr6_-_3157760 | 1.69 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chrX_-_45629661 | 1.61 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr1_+_144339738 | 1.60 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr3_+_190333097 | 1.58 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr7_-_122339162 | 1.53 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr16_+_56691606 | 1.48 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr15_-_72523454 | 1.47 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr21_-_28217721 | 1.44 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr9_-_117568365 | 1.43 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr5_-_16742330 | 1.39 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr10_-_74856608 | 1.37 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr22_+_39916558 | 1.35 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr8_-_119964434 | 1.35 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr6_+_31554779 | 1.33 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr1_-_89357179 | 1.31 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr1_-_165324983 | 1.30 |
ENST00000367893.4
|
LMX1A
|
LIM homeobox transcription factor 1, alpha |
| chr19_+_572543 | 1.29 |
ENST00000333511.3
ENST00000573216.1 ENST00000353555.4 |
BSG
|
basigin (Ok blood group) |
| chr17_-_79792909 | 1.28 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr17_-_39306054 | 1.27 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr16_+_56598961 | 1.25 |
ENST00000219162.3
|
MT4
|
metallothionein 4 |
| chr6_-_27835357 | 1.25 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr10_+_91152303 | 1.24 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr16_+_78056412 | 1.23 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr12_-_58165870 | 1.21 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr6_+_64282447 | 1.21 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_+_114256661 | 1.20 |
ENST00000306507.5
|
FOXD4L1
|
forkhead box D4-like 1 |
| chr17_+_32683456 | 1.19 |
ENST00000225844.2
|
CCL13
|
chemokine (C-C motif) ligand 13 |
| chr1_+_152881014 | 1.17 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr2_-_75426826 | 1.17 |
ENST00000305249.5
|
TACR1
|
tachykinin receptor 1 |
| chr11_-_61735103 | 1.16 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr11_+_62623621 | 1.12 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr10_-_90712520 | 1.11 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr12_-_99548270 | 1.10 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_159291090 | 1.10 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr16_+_56691911 | 1.10 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr17_-_48546232 | 1.08 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr17_-_39280419 | 1.06 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr11_-_615570 | 1.05 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr8_+_120428546 | 1.05 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr11_-_9025541 | 1.04 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr10_+_96522361 | 1.04 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr1_+_24069952 | 1.04 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr9_-_118417 | 1.01 |
ENST00000382500.2
|
FOXD4
|
forkhead box D4 |
| chr6_+_64281906 | 1.00 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chrX_+_153448107 | 0.97 |
ENST00000369935.5
|
OPN1MW
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chr15_+_85525205 | 0.95 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chr10_+_118305435 | 0.92 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr9_-_69202204 | 0.91 |
ENST00000377473.1
|
FOXD4L6
|
forkhead box D4-like 6 |
| chr1_-_153518270 | 0.91 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr10_-_95209 | 0.91 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr6_+_159290917 | 0.91 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr6_-_33771757 | 0.90 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr17_-_38911580 | 0.90 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr11_+_62623544 | 0.89 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_+_120254510 | 0.89 |
ENST00000369409.4
|
PHGDH
|
phosphoglycerate dehydrogenase |
| chr1_-_160313025 | 0.89 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr17_-_33700626 | 0.89 |
ENST00000308377.4
ENST00000394566.1 ENST00000589811.1 ENST00000430814.1 ENST00000427966.1 ENST00000588579.1 ENST00000591682.1 ENST00000441608.2 ENST00000592122.1 |
SLFN11
|
schlafen family member 11 |
| chr17_+_56315936 | 0.88 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr15_+_76629064 | 0.88 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr17_-_48546324 | 0.87 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr1_+_6105974 | 0.87 |
ENST00000378083.3
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_-_4535233 | 0.86 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr12_-_53074182 | 0.86 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr10_+_7745303 | 0.85 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_+_56270084 | 0.85 |
ENST00000225371.5
|
EPX
|
eosinophil peroxidase |
| chr6_+_27861190 | 0.84 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr11_+_62623512 | 0.82 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_+_13049413 | 0.81 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr6_+_31554612 | 0.81 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr11_+_1074870 | 0.81 |
ENST00000441003.2
ENST00000359061.5 |
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr22_+_32340447 | 0.80 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr7_+_29519662 | 0.80 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr19_-_36247910 | 0.79 |
ENST00000587965.1
ENST00000004982.3 |
HSPB6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr8_+_50822344 | 0.78 |
ENST00000518864.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr1_-_150552006 | 0.78 |
ENST00000307940.3
ENST00000369026.2 |
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related) |
| chr6_-_26124138 | 0.78 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr12_+_130646999 | 0.77 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr1_-_935491 | 0.77 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr11_-_36531774 | 0.77 |
ENST00000348124.5
ENST00000526995.1 |
TRAF6
|
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
| chr4_+_146539415 | 0.77 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr6_+_27107053 | 0.77 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr7_+_107531580 | 0.77 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr11_+_68451943 | 0.76 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr1_-_935361 | 0.76 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr22_+_32340481 | 0.76 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr18_-_49557 | 0.75 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr2_+_85766280 | 0.74 |
ENST00000306434.3
|
MAT2A
|
methionine adenosyltransferase II, alpha |
| chr16_+_2880369 | 0.74 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B |
| chr4_+_169842707 | 0.73 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_+_173116225 | 0.73 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr3_-_190167571 | 0.73 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr1_-_27481401 | 0.72 |
ENST00000263980.3
|
SLC9A1
|
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr21_+_19617140 | 0.72 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr17_-_39197699 | 0.71 |
ENST00000306271.4
|
KRTAP1-1
|
keratin associated protein 1-1 |
| chr11_+_60970852 | 0.71 |
ENST00000325558.6
|
PGA3
|
pepsinogen 3, group I (pepsinogen A) |
| chr17_+_80186908 | 0.70 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_-_141338627 | 0.70 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr2_-_75426183 | 0.70 |
ENST00000409848.3
|
TACR1
|
tachykinin receptor 1 |
| chr17_-_8113542 | 0.70 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr6_+_26156551 | 0.69 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_+_26204825 | 0.69 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr7_-_29186008 | 0.69 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr12_-_11508520 | 0.69 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr9_+_70917276 | 0.69 |
ENST00000342833.2
|
FOXD4L3
|
forkhead box D4-like 3 |
| chr4_-_70080449 | 0.68 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr7_+_72742178 | 0.68 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr10_-_69455873 | 0.68 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr17_+_76183432 | 0.68 |
ENST00000591256.1
ENST00000589256.1 ENST00000588800.1 ENST00000591952.1 ENST00000327898.5 ENST00000586542.1 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr17_+_56315787 | 0.68 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr20_+_18488528 | 0.67 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr21_-_19775973 | 0.67 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr15_+_41624892 | 0.67 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chrX_+_18725758 | 0.67 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr3_-_129035120 | 0.67 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr17_+_29248918 | 0.67 |
ENST00000581548.1
ENST00000580525.1 |
ADAP2
|
ArfGAP with dual PH domains 2 |
| chr18_+_9102592 | 0.66 |
ENST00000318388.6
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr12_-_81763127 | 0.66 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr17_+_32597232 | 0.66 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr12_+_6309517 | 0.66 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr7_+_143013198 | 0.66 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_-_27100529 | 0.65 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr11_-_61735029 | 0.65 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr11_+_67183557 | 0.65 |
ENST00000445895.2
|
CARNS1
|
carnosine synthase 1 |
| chr17_-_79849438 | 0.65 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr1_+_153330322 | 0.65 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr6_+_36562132 | 0.64 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr10_+_7745232 | 0.64 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_-_182360918 | 0.64 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr17_-_10325261 | 0.63 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chrX_-_48814810 | 0.63 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr7_+_116593292 | 0.63 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chrX_-_63615297 | 0.63 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr11_-_5255696 | 0.62 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr7_+_72742162 | 0.62 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr7_-_72439997 | 0.62 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr7_-_134143841 | 0.61 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr2_-_219858123 | 0.61 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chrX_+_149887090 | 0.61 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr9_+_140135665 | 0.61 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chrX_+_41548259 | 0.61 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr6_-_133035185 | 0.60 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr17_-_73150599 | 0.60 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr15_-_82939157 | 0.60 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr1_+_55446465 | 0.60 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chrX_+_41548220 | 0.59 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr17_-_76183111 | 0.59 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr18_+_56887381 | 0.59 |
ENST00000256857.2
ENST00000529320.2 ENST00000420468.2 |
GRP
|
gastrin-releasing peptide |
| chr1_-_152332480 | 0.58 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBP
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 3.0 | 12.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 1.4 | 4.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 1.3 | 3.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.8 | 3.8 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.7 | 2.2 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.7 | 17.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.7 | 2.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.7 | 1.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.6 | 1.8 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.6 | 2.4 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.6 | 2.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.5 | 14.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.5 | 16.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.5 | 2.5 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.5 | 2.0 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.5 | 2.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.4 | 1.3 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.4 | 2.1 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.4 | 1.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.4 | 1.2 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.4 | 1.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.3 | 2.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 2.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 0.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 0.9 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 0.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.3 | 2.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 3.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 0.8 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.2 | 0.7 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 0.7 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.2 | 3.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.9 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 6.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 1.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.2 | 0.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 0.6 | GO:0072061 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.2 | 0.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 0.8 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 0.8 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 5.0 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 0.8 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.6 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 0.9 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.9 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 0.5 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.2 | 0.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 2.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 0.6 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.6 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.7 | GO:0044878 | histone H3-S28 phosphorylation(GO:0043988) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 1.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 8.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 1.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.7 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 1.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 1.4 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 7.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.5 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.9 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 1.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.1 | 1.6 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 2.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 2.0 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.4 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 1.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0042322 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 1.1 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.8 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.7 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 1.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 2.6 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 2.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:2000302 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 1.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 4.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0044753 | amphisome(GO:0044753) |
| 0.5 | 2.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.5 | 5.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 2.2 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 1.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 2.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 2.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 0.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 0.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 1.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 2.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 12.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 3.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 15.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 2.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 7.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 3.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 3.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 2.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 15.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 10.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 6.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.9 | 14.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 18.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.5 | 4.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 2.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 2.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.9 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.4 | 2.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.4 | 1.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 0.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 1.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 2.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.6 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.2 | 1.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 0.6 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 2.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.5 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.2 | 5.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.8 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.5 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 8.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 11.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.8 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.5 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.8 | GO:0015266 | protein channel activity(GO:0015266) BH3 domain binding(GO:0051434) |
| 0.1 | 9.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 1.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.1 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 2.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.2 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 1.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 5.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 13.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 40.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 1.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 30.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 11.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 38.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 7.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 14.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 4.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.6 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 1.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 2.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |