Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
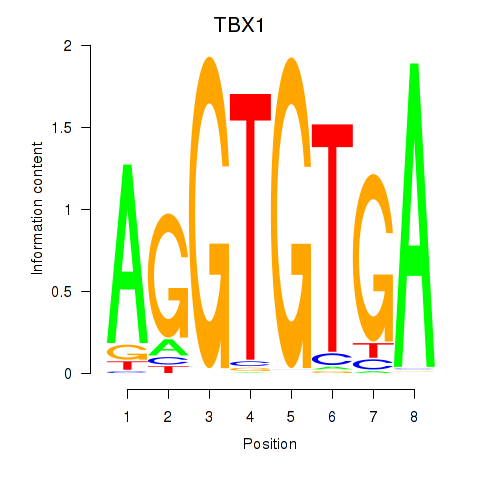
Results for TBX1
Z-value: 0.67
Transcription factors associated with TBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX1
|
ENSG00000184058.8 | T-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX1 | hg19_v2_chr22_+_19744226_19744226 | 0.24 | 2.5e-01 | Click! |
Activity profile of TBX1 motif
Sorted Z-values of TBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35454953 | 0.91 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr10_-_131762105 | 0.91 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr19_-_58864848 | 0.82 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr7_-_138363824 | 0.81 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr1_-_111174054 | 0.76 |
ENST00000369770.3
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr19_-_51875894 | 0.76 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr1_-_154127518 | 0.74 |
ENST00000368559.3
ENST00000271854.3 |
NUP210L
|
nucleoporin 210kDa-like |
| chr11_-_33913708 | 0.72 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_13487784 | 0.71 |
ENST00000379287.3
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr18_+_8717369 | 0.66 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr21_-_31869451 | 0.60 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr8_-_93107443 | 0.59 |
ENST00000360348.2
ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_+_100111479 | 0.58 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr9_-_74675521 | 0.56 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr17_+_68165657 | 0.54 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr1_-_238054094 | 0.50 |
ENST00000366570.4
|
ZP4
|
zona pellucida glycoprotein 4 |
| chr11_-_19262486 | 0.50 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr8_+_120220561 | 0.50 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr1_+_100111580 | 0.50 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr12_+_53443680 | 0.50 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr4_-_186732048 | 0.50 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_58212487 | 0.49 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr12_-_10022735 | 0.49 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr7_+_100663353 | 0.49 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chr19_-_13213662 | 0.49 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr11_+_103907308 | 0.48 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr1_-_153066998 | 0.47 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr19_+_37342547 | 0.46 |
ENST00000331800.4
ENST00000586646.1 |
ZNF345
|
zinc finger protein 345 |
| chr12_-_54691668 | 0.46 |
ENST00000553198.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_+_57810198 | 0.46 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr3_-_39234074 | 0.46 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr19_+_45445524 | 0.46 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr21_-_31859755 | 0.45 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr12_+_27396901 | 0.45 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr8_+_1993173 | 0.44 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr17_-_34122596 | 0.44 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr14_-_106733624 | 0.42 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr19_+_10527449 | 0.42 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr3_-_58523010 | 0.42 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr17_+_56769924 | 0.42 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr10_+_88414298 | 0.42 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr12_+_53443963 | 0.41 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr5_-_137514333 | 0.41 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr19_+_15904761 | 0.41 |
ENST00000308940.8
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5 |
| chr2_+_120687335 | 0.41 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr8_-_144099795 | 0.40 |
ENST00000522060.1
ENST00000517833.1 ENST00000502167.2 ENST00000518831.1 |
RP11-273G15.2
|
RP11-273G15.2 |
| chr19_+_42212501 | 0.40 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr7_+_140373619 | 0.40 |
ENST00000483369.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr5_+_68788594 | 0.40 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr22_+_40441456 | 0.40 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr7_-_99527243 | 0.39 |
ENST00000312891.2
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa |
| chr13_-_46543805 | 0.39 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr6_+_84743436 | 0.39 |
ENST00000257776.4
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr22_-_17073700 | 0.39 |
ENST00000359963.3
|
CCT8L2
|
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr10_-_102089729 | 0.39 |
ENST00000465680.2
|
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr10_-_46970474 | 0.39 |
ENST00000503753.1
ENST00000374321.4 |
SYT15
|
synaptotagmin XV |
| chr15_+_66585555 | 0.38 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr18_+_3466248 | 0.37 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr7_-_99149715 | 0.37 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr4_+_126237554 | 0.37 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr10_+_88414338 | 0.37 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr5_-_34043310 | 0.37 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr7_+_12726474 | 0.36 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr9_+_75136717 | 0.36 |
ENST00000297784.5
|
TMC1
|
transmembrane channel-like 1 |
| chr16_-_28550348 | 0.36 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr21_+_46117087 | 0.36 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr2_-_169769787 | 0.36 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr7_-_38398721 | 0.36 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3 |
| chr3_-_168865522 | 0.36 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_+_38893208 | 0.35 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr10_-_7513904 | 0.35 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr9_-_6015607 | 0.35 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr19_-_48752812 | 0.35 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr13_-_52027134 | 0.35 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr19_+_35417844 | 0.35 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr2_-_75796837 | 0.34 |
ENST00000233712.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr7_-_38403077 | 0.34 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr16_-_75282088 | 0.34 |
ENST00000542031.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr3_-_128212016 | 0.34 |
ENST00000498200.1
ENST00000341105.2 |
GATA2
|
GATA binding protein 2 |
| chr5_-_10308125 | 0.34 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr8_-_93029865 | 0.34 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_+_61547894 | 0.33 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr4_-_90757364 | 0.33 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_38017264 | 0.33 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_12058961 | 0.33 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr11_+_117947782 | 0.33 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr19_-_18995029 | 0.32 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr16_+_12059050 | 0.32 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_+_12250833 | 0.32 |
ENST00000396668.3
|
TMEM106B
|
transmembrane protein 106B |
| chr7_-_122635754 | 0.32 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr19_+_35417798 | 0.31 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr19_-_55458860 | 0.31 |
ENST00000592784.1
ENST00000448121.2 ENST00000340844.2 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr6_-_84419101 | 0.31 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr7_+_141490017 | 0.31 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr9_+_77230499 | 0.31 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr1_+_38022513 | 0.31 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr14_-_98444438 | 0.31 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr11_+_1940786 | 0.31 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr2_+_204571375 | 0.30 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr16_-_28550320 | 0.30 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr19_+_11909329 | 0.30 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr17_-_5026397 | 0.30 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr20_-_23433494 | 0.30 |
ENST00000377007.3
|
CST11
|
cystatin 11 |
| chr17_-_60005365 | 0.30 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr9_-_14314518 | 0.29 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr11_+_66824346 | 0.29 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr14_-_106453155 | 0.29 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr15_+_41913690 | 0.29 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr1_+_38022572 | 0.29 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chrX_-_52260199 | 0.29 |
ENST00000375600.1
|
XAGE1A
|
X antigen family, member 1A |
| chr22_-_32341336 | 0.29 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr12_-_1920886 | 0.29 |
ENST00000536846.2
ENST00000538027.2 ENST00000538450.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr2_+_234600253 | 0.29 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_241564655 | 0.29 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr12_-_3862245 | 0.29 |
ENST00000252322.1
ENST00000440314.2 |
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr19_+_859425 | 0.29 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr9_+_2157655 | 0.29 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_84873913 | 0.28 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease II beta |
| chr13_+_53602894 | 0.28 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chrX_-_63005405 | 0.28 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_+_44087729 | 0.28 |
ENST00000524990.1
ENST00000263776.8 |
ACCS
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr4_-_102268484 | 0.28 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_+_34680620 | 0.28 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr4_-_7941596 | 0.28 |
ENST00000420658.1
ENST00000358461.2 |
AFAP1
|
actin filament associated protein 1 |
| chr12_+_109785708 | 0.28 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr6_-_84937314 | 0.28 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr11_+_6260298 | 0.28 |
ENST00000379936.2
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr12_-_7848364 | 0.28 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr11_+_117947724 | 0.28 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_+_116184566 | 0.28 |
ENST00000355485.2
ENST00000369510.4 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chr13_+_50656307 | 0.28 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr12_-_68696652 | 0.28 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr5_-_72861484 | 0.28 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr9_+_40028620 | 0.28 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr2_+_233404429 | 0.27 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr13_+_114567131 | 0.27 |
ENST00000608651.1
|
GAS6-AS2
|
GAS6 antisense RNA 2 (head to head) |
| chr6_-_2842087 | 0.27 |
ENST00000537185.1
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr8_+_1993152 | 0.27 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr13_-_103019744 | 0.27 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr10_+_70320413 | 0.27 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr14_+_75746781 | 0.27 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr6_+_167738569 | 0.27 |
ENST00000239587.5
|
TTLL2
|
tubulin tyrosine ligase-like family, member 2 |
| chr1_+_27114418 | 0.27 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr8_+_104831554 | 0.27 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_15735925 | 0.27 |
ENST00000427824.1
|
RP3-467K16.4
|
RP3-467K16.4 |
| chr1_-_156786530 | 0.27 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr13_-_99404875 | 0.27 |
ENST00000376503.5
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr1_+_104068562 | 0.27 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_-_205744205 | 0.26 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr20_+_59654146 | 0.26 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr7_-_138386097 | 0.26 |
ENST00000421622.1
|
SVOPL
|
SVOP-like |
| chr16_-_88772761 | 0.26 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr7_+_12250886 | 0.26 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr2_-_219031709 | 0.26 |
ENST00000295683.2
|
CXCR1
|
chemokine (C-X-C motif) receptor 1 |
| chr9_+_140032842 | 0.26 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr19_+_37407212 | 0.26 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr7_-_27135591 | 0.26 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr19_+_35225060 | 0.26 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr17_-_7120525 | 0.26 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr17_-_7080227 | 0.26 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr9_-_14314566 | 0.26 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr19_+_18747775 | 0.26 |
ENST00000300976.4
ENST00000595182.1 ENST00000599006.1 |
KLHL26
|
kelch-like family member 26 |
| chr8_-_93107827 | 0.26 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr20_-_36661826 | 0.26 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr10_-_134331695 | 0.25 |
ENST00000455414.1
|
RP11-432J24.5
|
RP11-432J24.5 |
| chr9_+_125281420 | 0.25 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr22_-_37915535 | 0.25 |
ENST00000403299.1
|
CARD10
|
caspase recruitment domain family, member 10 |
| chr17_-_7123021 | 0.25 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr1_-_156786634 | 0.25 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr3_+_142315225 | 0.25 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr19_-_44405941 | 0.25 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr10_+_135160844 | 0.25 |
ENST00000423766.1
ENST00000458230.1 |
PRAP1
|
proline-rich acidic protein 1 |
| chr19_-_55574538 | 0.25 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr1_-_76076793 | 0.25 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr4_-_102268628 | 0.24 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr4_-_89619386 | 0.24 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr11_+_1940925 | 0.24 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr6_+_79577189 | 0.24 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr6_-_29324054 | 0.24 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr6_+_160221293 | 0.24 |
ENST00000610273.1
ENST00000392167.3 |
PNLDC1
|
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr17_+_53343577 | 0.24 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr1_+_89829610 | 0.24 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr6_+_127588020 | 0.24 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr19_-_22605136 | 0.24 |
ENST00000357774.5
ENST00000601553.1 ENST00000593657.1 |
ZNF98
|
zinc finger protein 98 |
| chr8_-_30891078 | 0.24 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr17_-_39968406 | 0.24 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr1_-_182641367 | 0.24 |
ENST00000508450.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr11_-_28129656 | 0.23 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr17_+_43922241 | 0.23 |
ENST00000329196.5
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr6_-_31560729 | 0.23 |
ENST00000340027.5
ENST00000376073.4 ENST00000376072.3 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr8_-_21999447 | 0.23 |
ENST00000306306.3
ENST00000521744.1 |
REEP4
|
receptor accessory protein 4 |
| chr5_+_64859066 | 0.23 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr19_+_46002868 | 0.23 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr15_-_74421477 | 0.23 |
ENST00000514871.1
|
RP11-247C2.2
|
HCG2004779; Uncharacterized protein |
| chr16_-_29479154 | 0.23 |
ENST00000549950.1
|
RP11-345J4.3
|
Uncharacterized protein |
| chr19_-_49540073 | 0.23 |
ENST00000301407.7
ENST00000601167.1 ENST00000604577.1 ENST00000591656.1 |
CGB1
CTB-60B18.6
|
chorionic gonadotropin, beta polypeptide 1 Choriogonadotropin subunit beta variant 1; Uncharacterized protein |
| chr12_+_32112340 | 0.23 |
ENST00000540924.1
ENST00000312561.4 |
KIAA1551
|
KIAA1551 |
| chr10_-_46970573 | 0.23 |
ENST00000374325.3
|
SYT15
|
synaptotagmin XV |
| chr2_-_264024 | 0.23 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr3_-_125094093 | 0.23 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.5 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.5 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.7 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.4 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.3 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.1 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.5 | GO:2000373 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.2 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.3 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.1 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.2 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.6 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0002086 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.5 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0045925 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:1901899 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0032764 | smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0090038 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0071106 | coenzyme A transport(GO:0015880) FAD transport(GO:0015883) coenzyme A transmembrane transport(GO:0035349) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:1904491 | somatic motor neuron differentiation(GO:0021523) protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0035568 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.0 | GO:0051884 | regulation of anagen(GO:0051884) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:1903464 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) negative regulation of mitotic cell cycle DNA replication(GO:1903464) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.0 | GO:0042097 | interleukin-10 biosynthetic process(GO:0042091) interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.5 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.4 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.4 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0051582 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.5 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.3 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.4 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.1 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.4 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.5 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.5 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.4 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.5 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.3 | GO:0035673 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) FAD transmembrane transporter activity(GO:0015230) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.0 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |