Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
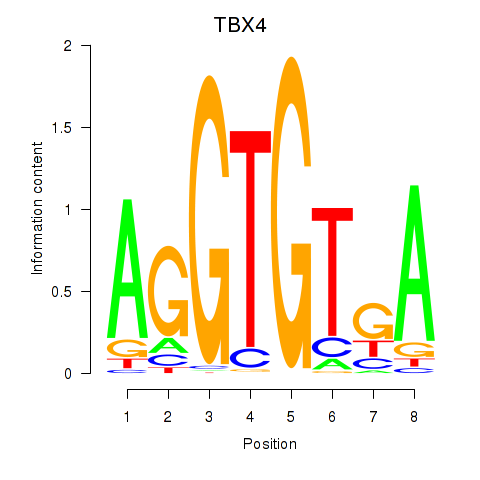
Results for TBX4
Z-value: 0.47
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.5 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX4 | hg19_v2_chr17_+_59529743_59529798 | -0.21 | 3.1e-01 | Click! |
Activity profile of TBX4 motif
Sorted Z-values of TBX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186732048 | 1.85 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_40028620 | 1.55 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr11_-_33913708 | 1.35 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr4_-_102268484 | 1.18 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_-_13487784 | 1.16 |
ENST00000379287.3
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr4_-_102268628 | 1.09 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_75567962 | 1.03 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr2_-_157189180 | 0.99 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr9_+_71986182 | 0.84 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_68788594 | 0.82 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr11_-_33891362 | 0.82 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr3_+_134514093 | 0.74 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr2_+_61108650 | 0.73 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr12_-_7848364 | 0.66 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr2_+_61108771 | 0.61 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr10_+_94833642 | 0.50 |
ENST00000224356.4
ENST00000394139.1 |
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr19_-_46526304 | 0.47 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr4_+_102268904 | 0.46 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr4_-_114682597 | 0.44 |
ENST00000394524.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr7_+_107220660 | 0.44 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr7_+_107220899 | 0.44 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr17_+_35849937 | 0.42 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr4_-_114682936 | 0.41 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr17_-_37557846 | 0.40 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr8_+_95907993 | 0.40 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_-_160473114 | 0.35 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_+_27114418 | 0.32 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr18_-_53068911 | 0.32 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr3_-_49459865 | 0.31 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr3_-_49459878 | 0.31 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr14_+_75746781 | 0.30 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chrX_-_133930285 | 0.29 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr13_+_115047097 | 0.29 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr11_-_18343669 | 0.29 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr2_+_62933001 | 0.28 |
ENST00000263991.5
ENST00000354487.3 |
EHBP1
|
EH domain binding protein 1 |
| chr16_-_2205352 | 0.28 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr1_+_154377669 | 0.27 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr2_+_103089756 | 0.27 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr10_+_45406627 | 0.26 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr20_+_11898507 | 0.26 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chrX_-_19689106 | 0.24 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr14_+_75745477 | 0.24 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr8_-_99954788 | 0.22 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr19_+_10131437 | 0.22 |
ENST00000587782.1
|
RDH8
|
retinol dehydrogenase 8 (all-trans) |
| chr2_-_160472952 | 0.22 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_+_62932779 | 0.21 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chrX_+_123095546 | 0.21 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr14_+_21467414 | 0.20 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr6_-_13487825 | 0.20 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr12_+_57853918 | 0.20 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr2_-_111291587 | 0.20 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr1_+_27114589 | 0.19 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr13_-_46543805 | 0.19 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr17_-_39203519 | 0.19 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr11_-_114271139 | 0.18 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr2_-_61697862 | 0.17 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr8_-_95907423 | 0.17 |
ENST00000396133.3
ENST00000308108.4 |
CCNE2
|
cyclin E2 |
| chrX_+_123480375 | 0.16 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr22_+_40441456 | 0.16 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr16_+_67596310 | 0.16 |
ENST00000264010.4
ENST00000401394.1 |
CTCF
|
CCCTC-binding factor (zinc finger protein) |
| chr12_+_104324112 | 0.15 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr12_-_90024360 | 0.15 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_+_89402148 | 0.14 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr14_+_23344345 | 0.14 |
ENST00000551466.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr9_-_74979420 | 0.14 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr8_+_103563792 | 0.14 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr1_+_145507587 | 0.14 |
ENST00000330165.8
ENST00000369307.3 |
RBM8A
|
RNA binding motif protein 8A |
| chrX_+_123480421 | 0.14 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr11_+_4116054 | 0.14 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr11_+_111749650 | 0.14 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr17_-_26220366 | 0.14 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr16_+_85645007 | 0.14 |
ENST00000405402.2
|
GSE1
|
Gse1 coiled-coil protein |
| chr7_-_127032741 | 0.13 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr13_-_52027134 | 0.13 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_+_18343800 | 0.13 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr6_+_32132360 | 0.13 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr12_-_91348949 | 0.13 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr15_-_90294523 | 0.13 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr5_-_74807418 | 0.12 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr1_+_228395755 | 0.12 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr4_-_140222358 | 0.12 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_+_39279838 | 0.11 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr14_-_45603657 | 0.11 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr12_+_50451331 | 0.11 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr2_-_85895295 | 0.11 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr1_-_44482979 | 0.11 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_27282319 | 0.11 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr1_-_1009683 | 0.10 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chrX_+_46771848 | 0.10 |
ENST00000218343.4
|
PHF16
|
jade family PHD finger 3 |
| chr6_+_79577189 | 0.10 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chrX_+_46771711 | 0.10 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chr10_+_7860460 | 0.10 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr11_-_7818520 | 0.10 |
ENST00000329434.2
|
OR5P2
|
olfactory receptor, family 5, subfamily P, member 2 |
| chr20_+_58630972 | 0.10 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr11_+_1940786 | 0.10 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr5_+_56469939 | 0.10 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_+_172864490 | 0.09 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr1_-_85930246 | 0.09 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_111750206 | 0.09 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr2_+_37458776 | 0.08 |
ENST00000002125.4
ENST00000336237.6 ENST00000431821.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr20_+_2517253 | 0.08 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr22_-_20231207 | 0.08 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr5_+_56469843 | 0.08 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr14_+_53196872 | 0.08 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr5_+_74807886 | 0.08 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr3_+_52811596 | 0.07 |
ENST00000542827.1
ENST00000273283.2 |
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr11_+_122709200 | 0.07 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr12_-_53097247 | 0.07 |
ENST00000341809.3
ENST00000537195.1 |
KRT77
|
keratin 77 |
| chr7_+_133261209 | 0.07 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr11_+_18344106 | 0.07 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr21_+_30502806 | 0.07 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_+_120081475 | 0.07 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr3_+_63805017 | 0.07 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr1_-_226076843 | 0.06 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr17_-_61973929 | 0.06 |
ENST00000329882.8
ENST00000453363.3 ENST00000316193.8 |
CSH1
|
chorionic somatomammotropin hormone 1 (placental lactogen) |
| chr5_+_74807581 | 0.06 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr14_+_90864504 | 0.05 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_+_16062820 | 0.05 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr6_+_39760129 | 0.05 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chrX_+_123480194 | 0.05 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr15_+_41913690 | 0.05 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr3_+_142315225 | 0.05 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr7_-_71801980 | 0.05 |
ENST00000329008.5
|
CALN1
|
calneuron 1 |
| chr12_+_21525818 | 0.05 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr11_-_129062093 | 0.05 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr5_+_56469775 | 0.04 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr8_+_73449625 | 0.04 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr9_-_74980113 | 0.04 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chrM_+_10053 | 0.04 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr15_-_83474806 | 0.04 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr11_+_111749942 | 0.03 |
ENST00000260276.3
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr17_+_30771279 | 0.03 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr5_-_54281491 | 0.03 |
ENST00000381405.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr2_-_40680578 | 0.03 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr4_-_186456766 | 0.03 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr11_-_118972575 | 0.02 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr12_+_65672702 | 0.02 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr1_+_114471972 | 0.02 |
ENST00000369559.4
ENST00000369554.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_-_37458749 | 0.02 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr16_-_67965756 | 0.02 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr11_-_5248294 | 0.02 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr11_-_34535332 | 0.01 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr3_+_137728842 | 0.01 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr13_-_36429763 | 0.01 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_-_186456652 | 0.01 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr11_+_114271251 | 0.01 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr8_+_87111059 | 0.01 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr5_-_60240858 | 0.01 |
ENST00000426742.2
ENST00000265038.5 ENST00000543101.1 ENST00000439176.1 |
ERCC8
|
excision repair cross-complementing rodent repair deficiency, complementation group 8 |
| chr18_-_56296182 | 0.01 |
ENST00000361673.3
|
ALPK2
|
alpha-kinase 2 |
| chr12_+_58148842 | 0.01 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr7_-_27142290 | 0.01 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr5_-_54281407 | 0.01 |
ENST00000381403.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr1_+_114471809 | 0.01 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr17_+_57970469 | 0.00 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 1.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 1.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.7 | GO:1902725 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.5 | GO:0032824 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.5 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.6 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.7 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.2 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 1.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.2 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.8 | GO:1901725 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) regulation of histone deacetylase activity(GO:1901725) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 1.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.8 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.2 | GO:0070602 | maintenance of DNA methylation(GO:0010216) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.4 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.6 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 2.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |