Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
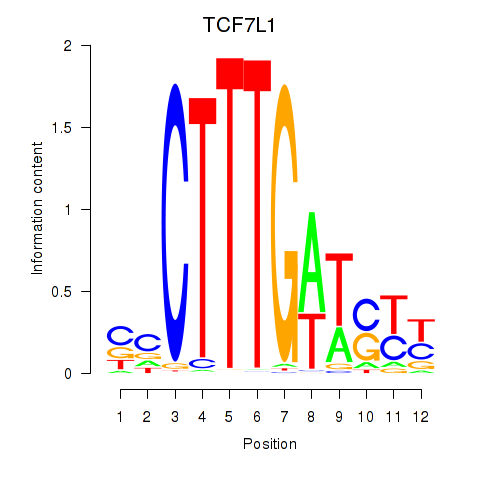
Results for TCF7L1
Z-value: 0.72
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | transcription factor 7 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L1 | hg19_v2_chr2_+_85360499_85360598 | -0.02 | 9.3e-01 | Click! |
Activity profile of TCF7L1 motif
Sorted Z-values of TCF7L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_72440901 | 2.68 |
ENST00000359684.2
|
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr13_-_72441315 | 2.65 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr4_-_186877502 | 2.19 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_61330931 | 1.77 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr9_+_75263565 | 1.11 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr2_+_69240302 | 1.09 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr1_+_82266053 | 0.89 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr3_-_148804275 | 0.86 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr7_-_27213893 | 0.86 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr3_+_20081515 | 0.84 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr10_+_54074033 | 0.71 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr5_+_175288631 | 0.71 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr14_+_21236586 | 0.68 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr14_+_75230011 | 0.65 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr2_+_69240511 | 0.65 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr17_+_30813576 | 0.63 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_+_16085244 | 0.61 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr10_+_112631547 | 0.60 |
ENST00000280154.7
ENST00000393104.2 |
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr10_-_25241499 | 0.60 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr20_+_30598231 | 0.59 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr19_-_11450249 | 0.59 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr16_+_30406721 | 0.57 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr5_+_170846640 | 0.55 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr2_+_11679963 | 0.54 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr7_-_27219849 | 0.51 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr14_+_23846328 | 0.50 |
ENST00000382809.2
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr2_+_16080659 | 0.49 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chrX_-_39956656 | 0.49 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr1_-_94079648 | 0.48 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_208031542 | 0.48 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr15_-_82338460 | 0.46 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr16_-_4850471 | 0.46 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr11_-_76155618 | 0.45 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr13_+_39612485 | 0.44 |
ENST00000379599.2
|
NHLRC3
|
NHL repeat containing 3 |
| chr11_-_76155700 | 0.44 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr12_+_56324933 | 0.43 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr3_+_123813509 | 0.42 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr2_+_69240415 | 0.42 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr11_-_62474803 | 0.42 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_+_71562180 | 0.41 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr13_+_39612442 | 0.41 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr4_-_157892498 | 0.41 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr14_+_75746781 | 0.39 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr9_+_116225999 | 0.38 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr12_+_56324756 | 0.38 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr4_-_157892167 | 0.37 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr13_+_76210448 | 0.36 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr8_+_29952914 | 0.36 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr17_+_80416050 | 0.35 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr21_+_18811205 | 0.33 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr17_+_80416482 | 0.33 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr17_-_46690839 | 0.32 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr11_-_87908600 | 0.32 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chrX_-_100183894 | 0.32 |
ENST00000328526.5
ENST00000372956.2 |
XKRX
|
XK, Kell blood group complex subunit-related, X-linked |
| chr6_+_96025341 | 0.31 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr7_+_115850547 | 0.31 |
ENST00000358204.4
ENST00000455989.1 ENST00000537767.1 |
TES
|
testis derived transcript (3 LIM domains) |
| chr16_-_15736881 | 0.31 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr3_+_52813932 | 0.31 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr12_+_80603233 | 0.30 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr10_+_71561704 | 0.30 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr10_+_115312766 | 0.30 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr12_+_9067123 | 0.29 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr10_+_76585303 | 0.29 |
ENST00000372725.1
|
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr20_-_17511962 | 0.28 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr7_+_73868439 | 0.28 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr10_+_71561630 | 0.28 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr1_+_59762642 | 0.28 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr8_+_29953163 | 0.28 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr6_+_143929307 | 0.28 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr8_-_16859690 | 0.27 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chrX_-_117119243 | 0.27 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr4_-_157892055 | 0.26 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr11_-_94965667 | 0.26 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr2_-_208031943 | 0.26 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_-_7297833 | 0.26 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chrX_-_32173579 | 0.25 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr17_+_35851570 | 0.25 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr10_+_71561649 | 0.25 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_179184955 | 0.25 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr14_+_105941118 | 0.24 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr4_-_70626314 | 0.24 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chrX_+_135229600 | 0.23 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr13_+_76334498 | 0.23 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr7_+_90339169 | 0.23 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr7_-_19813192 | 0.22 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr15_-_64995399 | 0.22 |
ENST00000559753.1
ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr1_+_93913713 | 0.22 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr16_-_4852915 | 0.22 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr17_-_7297519 | 0.22 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr10_-_87551311 | 0.22 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr5_-_175964366 | 0.22 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr2_+_234621551 | 0.22 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr3_+_137483579 | 0.22 |
ENST00000306087.1
|
SOX14
|
SRY (sex determining region Y)-box 14 |
| chr7_+_80275621 | 0.21 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr6_-_32145861 | 0.21 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr4_+_113152978 | 0.21 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr16_-_73082274 | 0.21 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chrX_+_135229559 | 0.21 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chr5_-_148930960 | 0.20 |
ENST00000261798.5
ENST00000377843.2 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chrX_-_135962876 | 0.20 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr14_-_35344093 | 0.20 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chrX_-_128788914 | 0.20 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr7_-_148581251 | 0.20 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr16_-_18812746 | 0.20 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr7_-_148581360 | 0.19 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr14_-_23624511 | 0.19 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr18_+_29769978 | 0.19 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr9_-_27005686 | 0.19 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr1_+_87794150 | 0.19 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr4_+_113152881 | 0.19 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_54303934 | 0.19 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr18_-_31802282 | 0.18 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr10_+_7745303 | 0.18 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr13_+_76334795 | 0.18 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr21_+_30671690 | 0.18 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr18_-_31802056 | 0.18 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr7_+_128828713 | 0.18 |
ENST00000249373.3
|
SMO
|
smoothened, frizzled family receptor |
| chr13_+_76334567 | 0.17 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr13_+_24144796 | 0.17 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr8_-_33424636 | 0.17 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chrX_+_100646190 | 0.17 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr6_+_119215308 | 0.17 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr20_+_18794370 | 0.17 |
ENST00000377428.2
|
SCP2D1
|
SCP2 sterol-binding domain containing 1 |
| chr10_+_22605374 | 0.17 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr19_-_48048518 | 0.17 |
ENST00000595558.1
ENST00000263351.5 |
ZNF541
|
zinc finger protein 541 |
| chr3_+_183353356 | 0.16 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr16_+_2820912 | 0.16 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr17_+_75447326 | 0.16 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr14_-_23451467 | 0.16 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr6_-_107436473 | 0.16 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr10_+_22605304 | 0.16 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr17_-_56065484 | 0.16 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr10_+_7745232 | 0.15 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_-_99052382 | 0.15 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr1_+_200993071 | 0.15 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr5_+_43603229 | 0.15 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chrX_+_152760397 | 0.15 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chr17_+_57297807 | 0.15 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr17_+_62223320 | 0.15 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr12_+_70760056 | 0.15 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr8_-_6914251 | 0.15 |
ENST00000330590.2
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific |
| chr12_-_48963829 | 0.14 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr10_+_11206925 | 0.14 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr4_+_78078304 | 0.14 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr8_-_29940464 | 0.14 |
ENST00000521265.1
ENST00000536273.1 |
TMEM66
|
transmembrane protein 66 |
| chr1_-_54303949 | 0.14 |
ENST00000234725.8
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr11_-_65381643 | 0.14 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr8_+_32579341 | 0.14 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr14_+_21214039 | 0.14 |
ENST00000326842.2
|
EDDM3A
|
epididymal protein 3A |
| chr1_+_36348790 | 0.14 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr12_+_53491220 | 0.14 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr7_+_80231466 | 0.14 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_104192405 | 0.13 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr6_+_32146131 | 0.13 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr7_+_80275752 | 0.13 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_+_52043758 | 0.13 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr19_+_11071546 | 0.13 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr18_+_3449821 | 0.13 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_11779403 | 0.12 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr15_-_79103757 | 0.12 |
ENST00000388820.4
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chrX_+_100645812 | 0.12 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chrX_-_153599578 | 0.12 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chrX_+_54835493 | 0.12 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr21_-_34144157 | 0.12 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr4_+_170581213 | 0.11 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr14_+_39644387 | 0.11 |
ENST00000553331.1
ENST00000216832.4 |
PNN
|
pinin, desmosome associated protein |
| chr20_+_55966444 | 0.11 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr13_+_24144509 | 0.11 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr4_+_79567314 | 0.11 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr4_-_85419603 | 0.11 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr1_+_154244987 | 0.10 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr14_-_73493784 | 0.10 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr1_+_212797789 | 0.10 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr19_+_8274204 | 0.10 |
ENST00000561053.1
ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4
|
ceramide synthase 4 |
| chr19_+_8274185 | 0.10 |
ENST00000558268.1
ENST00000558331.1 |
CERS4
|
ceramide synthase 4 |
| chr1_-_20126365 | 0.10 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr2_+_48796120 | 0.10 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr2_+_33172221 | 0.10 |
ENST00000354476.3
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr19_+_11071652 | 0.10 |
ENST00000344626.4
ENST00000429416.3 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr1_+_78383813 | 0.10 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr8_-_145047688 | 0.10 |
ENST00000356346.3
|
PLEC
|
plectin |
| chr4_+_79567362 | 0.10 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr7_+_90338712 | 0.09 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr17_-_61819229 | 0.09 |
ENST00000447001.3
ENST00000392950.4 |
STRADA
|
STE20-related kinase adaptor alpha |
| chr14_-_23451845 | 0.09 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr17_-_61819121 | 0.09 |
ENST00000245865.5
ENST00000375840.4 ENST00000582137.1 ENST00000579549.1 ENST00000582030.1 ENST00000584110.1 ENST00000580288.1 ENST00000336174.6 ENST00000579340.1 ENST00000580338.1 |
STRADA
|
STE20-related kinase adaptor alpha |
| chr9_-_74979420 | 0.09 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr10_-_91295304 | 0.08 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr7_+_80275953 | 0.08 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_185703513 | 0.08 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr14_+_20923350 | 0.08 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr10_-_104179682 | 0.08 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr19_-_49864746 | 0.07 |
ENST00000598810.1
|
TEAD2
|
TEA domain family member 2 |
| chr22_+_24105394 | 0.07 |
ENST00000305199.5
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr1_-_54304212 | 0.07 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr3_-_149375783 | 0.07 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr9_-_35650900 | 0.07 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr5_-_114515734 | 0.07 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr3_-_134093275 | 0.07 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr13_-_67802549 | 0.07 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.7 | 5.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 0.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.6 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 0.6 | GO:0036446 | myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.2 | 0.6 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 1.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.4 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.4 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.6 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.6 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 2.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.2 | GO:1904020 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 1.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.2 | GO:0007418 | positive regulation of hh target transcription factor activity(GO:0007228) ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 1.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 1.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.7 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 6.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |