Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
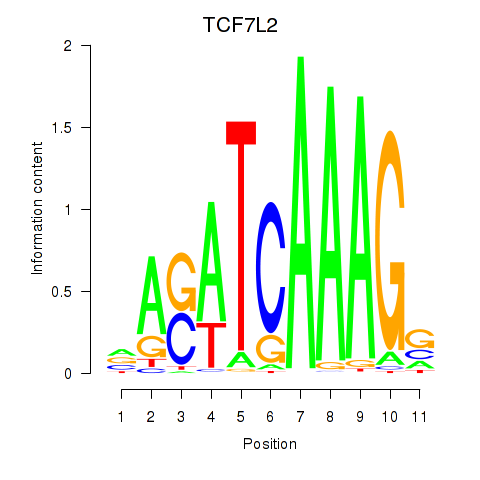
Results for TCF7L2
Z-value: 0.47
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.11 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg19_v2_chr10_+_114710516_114710675 | -0.19 | 3.7e-01 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_72441315 | 1.62 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr13_-_72440901 | 1.58 |
ENST00000359684.2
|
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr1_+_61330931 | 1.34 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr7_-_27213893 | 1.12 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr7_-_27219849 | 0.89 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr11_+_7597639 | 0.78 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chrX_-_63005405 | 0.75 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_+_69240302 | 0.65 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr13_+_24144796 | 0.64 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_162351503 | 0.58 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr17_-_67138015 | 0.58 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr10_+_54074033 | 0.53 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_+_29952914 | 0.52 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr5_-_111093167 | 0.49 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr2_+_89890533 | 0.44 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr4_+_30721968 | 0.41 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr8_+_29953163 | 0.40 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr13_+_24144509 | 0.39 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr5_+_175288631 | 0.37 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr22_-_32860427 | 0.37 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chr13_-_67802549 | 0.37 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr8_-_16859690 | 0.35 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr13_+_76334498 | 0.35 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr20_-_17511962 | 0.35 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr1_-_110933663 | 0.33 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr7_-_148581360 | 0.33 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chrX_-_10588595 | 0.33 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr18_+_46065483 | 0.31 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chrX_-_10588459 | 0.31 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr1_-_110933611 | 0.30 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr1_-_94079648 | 0.30 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_25241499 | 0.30 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr7_-_148581251 | 0.30 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chrX_-_32173579 | 0.28 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr21_+_18811205 | 0.28 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr16_+_29819372 | 0.28 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_29819446 | 0.27 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_+_45944190 | 0.26 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr2_+_177028805 | 0.26 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr12_-_48963829 | 0.26 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr5_-_148930960 | 0.25 |
ENST00000261798.5
ENST00000377843.2 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr2_+_69240511 | 0.24 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr1_-_161993616 | 0.24 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr12_-_12491608 | 0.24 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr5_-_135290705 | 0.24 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr18_+_46065393 | 0.24 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr7_+_27282319 | 0.24 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr7_+_99613195 | 0.23 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr6_-_32145861 | 0.23 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr12_+_9067123 | 0.22 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr15_+_57511609 | 0.22 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr2_-_55647057 | 0.22 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_-_55646957 | 0.22 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr11_-_94965667 | 0.22 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr13_+_76334567 | 0.22 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr12_-_54694807 | 0.22 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr10_+_101542462 | 0.22 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr11_-_76155618 | 0.21 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr13_+_76334795 | 0.21 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr1_+_147013182 | 0.21 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr7_+_73868439 | 0.21 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_+_78383813 | 0.21 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr14_+_75230011 | 0.20 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr11_-_62474803 | 0.20 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_40315640 | 0.20 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr15_-_90294523 | 0.20 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr12_+_80603233 | 0.20 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr18_+_29769978 | 0.20 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr11_-_76155700 | 0.19 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_63557759 | 0.19 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr2_+_173686303 | 0.19 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_69240415 | 0.19 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr14_-_81425828 | 0.18 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr2_+_204801471 | 0.18 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr1_-_40367668 | 0.17 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr19_+_50084561 | 0.17 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr22_+_19950060 | 0.17 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chr20_+_59654146 | 0.16 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr11_-_41481135 | 0.16 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chrX_+_54835493 | 0.16 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr6_-_112575838 | 0.16 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr7_-_80141328 | 0.15 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr17_-_38256973 | 0.15 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr3_-_134093275 | 0.14 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr17_+_68100989 | 0.14 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_54303934 | 0.14 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr12_+_70760056 | 0.14 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr9_+_125796806 | 0.14 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chrX_-_100183894 | 0.14 |
ENST00000328526.5
ENST00000372956.2 |
XKRX
|
XK, Kell blood group complex subunit-related, X-linked |
| chr3_+_183353356 | 0.14 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr17_+_75447326 | 0.14 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr1_+_207039154 | 0.14 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr6_-_28220002 | 0.14 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr16_-_73082274 | 0.14 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr2_+_33701286 | 0.14 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_48796120 | 0.13 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr17_+_79679299 | 0.13 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr7_+_80275621 | 0.13 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chrX_+_152760397 | 0.13 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chr6_+_155537771 | 0.13 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_-_112575758 | 0.13 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr1_+_223101757 | 0.13 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr9_-_130712995 | 0.13 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr17_+_42081914 | 0.13 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr6_+_32146131 | 0.13 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr2_+_234621551 | 0.12 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr10_-_105677427 | 0.12 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr12_+_85673868 | 0.12 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr2_+_201936458 | 0.12 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr3_-_134093395 | 0.12 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr9_-_15510287 | 0.12 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chrX_-_110655306 | 0.12 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr3_-_149375783 | 0.11 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr4_-_69215467 | 0.11 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1 |
| chr9_+_139221880 | 0.11 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr4_+_170581213 | 0.11 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr1_-_109935819 | 0.11 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr8_-_133123406 | 0.11 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr12_+_6982725 | 0.11 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chrX_+_46937745 | 0.10 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chrX_+_41306575 | 0.10 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr6_-_112575687 | 0.10 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr10_-_104179682 | 0.10 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_+_154244987 | 0.10 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr7_+_99613212 | 0.10 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_-_54304212 | 0.09 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr5_+_102201509 | 0.09 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_21236586 | 0.09 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr5_+_102201430 | 0.09 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_-_6982442 | 0.09 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chrX_+_106163626 | 0.09 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr17_+_80416050 | 0.09 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr3_-_33686743 | 0.09 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr20_+_57875658 | 0.09 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr1_+_180199393 | 0.09 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr18_-_52989217 | 0.09 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr5_+_60628074 | 0.09 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr4_+_78078304 | 0.09 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chrX_+_47004599 | 0.09 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr14_+_23235886 | 0.09 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr6_-_112575912 | 0.09 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr2_+_201936707 | 0.08 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr1_-_54303949 | 0.08 |
ENST00000234725.8
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr1_+_16330723 | 0.08 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
| chr17_+_80416482 | 0.08 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr6_+_152130240 | 0.08 |
ENST00000427531.2
|
ESR1
|
estrogen receptor 1 |
| chr7_+_150076406 | 0.08 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr10_-_87551311 | 0.07 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr3_+_174577070 | 0.07 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr10_-_104192405 | 0.07 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr13_-_36429763 | 0.07 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chrX_-_39956656 | 0.07 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr16_+_53242350 | 0.07 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_166028709 | 0.07 |
ENST00000595430.1
|
AL626787.1
|
AL626787.1 |
| chr17_-_39646116 | 0.07 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr12_-_52779433 | 0.07 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr3_-_42452050 | 0.06 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chr15_-_82338460 | 0.06 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr4_-_157892498 | 0.06 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr10_+_71561704 | 0.06 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_+_34848049 | 0.06 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr2_+_71357744 | 0.06 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_132873832 | 0.06 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chrX_+_47004639 | 0.06 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr1_-_115053781 | 0.06 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr17_-_47308128 | 0.06 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chrX_-_153599578 | 0.05 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr16_+_72088376 | 0.05 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr19_-_59030921 | 0.05 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr19_-_59031118 | 0.05 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr5_-_157002775 | 0.05 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr20_+_57875457 | 0.05 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr15_-_89764929 | 0.05 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chrX_+_100646190 | 0.05 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr17_-_7297833 | 0.04 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr8_-_95220775 | 0.04 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_-_7297519 | 0.04 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr14_-_75179774 | 0.04 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr5_+_54455946 | 0.04 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr7_+_80275752 | 0.04 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr22_+_41347363 | 0.04 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr4_-_157892055 | 0.04 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr11_-_6677018 | 0.04 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr20_+_57875758 | 0.04 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr10_+_71561630 | 0.03 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_-_109684120 | 0.03 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr3_+_137483579 | 0.03 |
ENST00000306087.1
|
SOX14
|
SRY (sex determining region Y)-box 14 |
| chr7_-_148725733 | 0.03 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr21_-_35284635 | 0.03 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr5_+_175815732 | 0.03 |
ENST00000274787.2
|
HIGD2A
|
HIG1 hypoxia inducible domain family, member 2A |
| chr12_+_52056548 | 0.03 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr17_-_40169659 | 0.03 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chrX_-_122866874 | 0.02 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr10_+_115312766 | 0.02 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chrX_+_100645812 | 0.02 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr2_+_204732487 | 0.02 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr17_-_47308100 | 0.02 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr19_-_39924349 | 0.02 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr7_+_110731062 | 0.01 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 1.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.6 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.1 | 2.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.4 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.2 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:1904833 | L-ascorbic acid biosynthetic process(GO:0019853) positive regulation of superoxide dismutase activity(GO:1901671) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.4 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |