Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
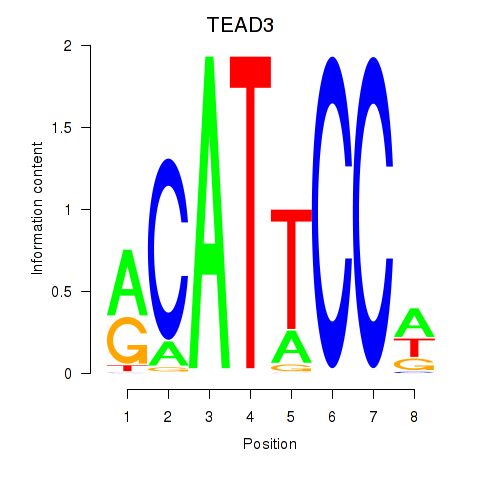
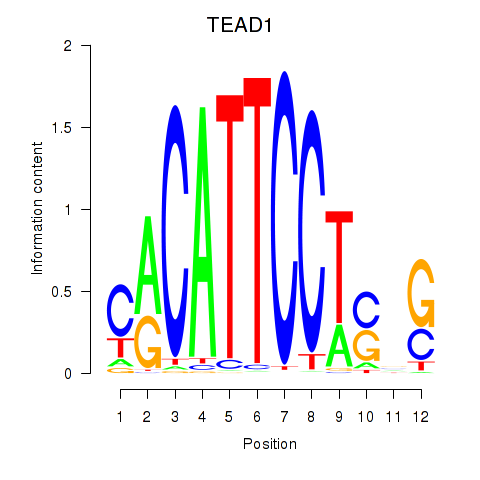
Results for TEAD3_TEAD1
Z-value: 1.47
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEA domain transcription factor 3 |
|
TEAD1
|
ENSG00000187079.10 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg19_v2_chr11_+_12695944_12695989 | 0.75 | 1.4e-05 | Click! |
| TEAD3 | hg19_v2_chr6_-_35464817_35464894, hg19_v2_chr6_-_35464727_35464738 | 0.13 | 5.3e-01 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186733363 | 7.73 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_39425068 | 7.00 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_+_41614720 | 6.66 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41614909 | 5.43 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr6_+_30850697 | 4.97 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_+_32605437 | 4.82 |
ENST00000380250.3
|
FRY
|
furry homolog (Drosophila) |
| chr12_-_96184533 | 4.04 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr8_+_144816303 | 3.58 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr4_-_186877806 | 3.53 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_14308004 | 3.26 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr6_-_132272504 | 3.16 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr5_-_39425222 | 2.97 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_186877502 | 2.97 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_39425290 | 2.78 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_+_34972543 | 2.78 |
ENST00000590071.2
|
WTIP
|
Wilms tumor 1 interacting protein |
| chr1_+_78354297 | 2.78 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr8_-_101322132 | 2.30 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_+_78354175 | 2.21 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr4_-_186732048 | 2.21 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_30960915 | 2.18 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr15_+_96869165 | 2.18 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_66024213 | 2.18 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr9_-_13175823 | 2.12 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr1_+_61548225 | 2.10 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr16_+_29823427 | 2.09 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr10_+_94590910 | 1.99 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr6_-_30712313 | 1.97 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr9_-_14314518 | 1.96 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr11_+_66824276 | 1.96 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr9_-_14314566 | 1.94 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr6_-_32191834 | 1.94 |
ENST00000375023.3
|
NOTCH4
|
notch 4 |
| chr1_-_59043166 | 1.93 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr11_+_66824346 | 1.83 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr5_-_55290773 | 1.77 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr1_+_61548374 | 1.70 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr9_+_82187487 | 1.70 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_2015335 | 1.68 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_2159850 | 1.62 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_114522049 | 1.60 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chrX_-_10588459 | 1.60 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr20_-_62680984 | 1.59 |
ENST00000340356.7
|
SOX18
|
SRY (sex determining region Y)-box 18 |
| chr1_-_94050668 | 1.57 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr6_-_41909191 | 1.55 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr15_+_39873268 | 1.54 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr3_-_149051444 | 1.53 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr9_+_82187630 | 1.52 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chrX_+_10031499 | 1.51 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr6_-_41909561 | 1.51 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr3_-_149051194 | 1.48 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr2_-_111230393 | 1.48 |
ENST00000447537.2
ENST00000413601.2 |
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr6_-_41909466 | 1.47 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr2_+_189157498 | 1.43 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_-_152149033 | 1.42 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr3_-_112360116 | 1.42 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr2_+_110656005 | 1.39 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr17_-_40575535 | 1.38 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr1_+_66999799 | 1.37 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr12_-_12419703 | 1.34 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr17_+_48911942 | 1.33 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr16_+_29823552 | 1.32 |
ENST00000300797.6
|
PRRT2
|
proline-rich transmembrane protein 2 |
| chr9_+_131314859 | 1.32 |
ENST00000358161.5
ENST00000372731.4 ENST00000372739.3 |
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 |
| chr10_-_33625154 | 1.29 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr12_-_104443890 | 1.19 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr20_-_23066953 | 1.19 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr21_+_17566643 | 1.18 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_7758374 | 1.16 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr12_+_27677085 | 1.16 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr4_-_70725856 | 1.12 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr3_-_114343039 | 1.12 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_46316677 | 1.11 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr2_+_109271481 | 1.10 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_-_38325219 | 1.10 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr20_+_42574317 | 1.09 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr11_+_58390132 | 1.08 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr2_-_75788038 | 1.08 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr1_+_230193521 | 1.07 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chrY_+_16634483 | 1.06 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr10_+_99344104 | 1.06 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr1_+_86046433 | 1.05 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr4_+_140586922 | 1.05 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr14_+_63671105 | 1.03 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr2_+_189157536 | 1.03 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_+_66511540 | 1.03 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr6_+_116832789 | 1.03 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr1_+_28764653 | 1.01 |
ENST00000373836.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr14_+_63671577 | 0.99 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr2_+_198669365 | 0.98 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr1_-_154474589 | 0.98 |
ENST00000304760.2
|
SHE
|
Src homology 2 domain containing E |
| chr9_+_102584128 | 0.97 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr2_+_106682135 | 0.97 |
ENST00000437659.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr1_+_100315613 | 0.96 |
ENST00000361915.3
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr12_+_32655048 | 0.96 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_106682103 | 0.94 |
ENST00000238044.3
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr7_-_27219849 | 0.93 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr18_-_53068911 | 0.93 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr19_+_45409011 | 0.93 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr21_-_40033618 | 0.93 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr18_-_53069419 | 0.92 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr19_+_39903185 | 0.92 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_-_119234876 | 0.92 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr6_-_111804905 | 0.91 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr17_-_39677971 | 0.91 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr7_+_155090271 | 0.91 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr2_+_45878790 | 0.89 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr6_+_143999072 | 0.88 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr14_-_23904861 | 0.86 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr6_+_148663729 | 0.86 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr2_+_170550944 | 0.85 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr17_-_67057047 | 0.85 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr17_-_67057114 | 0.84 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr9_+_27109392 | 0.84 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr2_-_183387430 | 0.83 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_95158375 | 0.83 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr8_+_97597148 | 0.82 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr8_-_81787006 | 0.82 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chrX_-_10851762 | 0.82 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_+_92085262 | 0.82 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr1_+_100818484 | 0.82 |
ENST00000544534.1
|
CDC14A
|
cell division cycle 14A |
| chr7_-_150754935 | 0.81 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr4_-_174255400 | 0.81 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr3_+_152017924 | 0.81 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr5_-_95158644 | 0.80 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr3_+_25469802 | 0.79 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr8_+_70378852 | 0.78 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr11_+_92577506 | 0.78 |
ENST00000533797.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr16_-_49698136 | 0.78 |
ENST00000535559.1
|
ZNF423
|
zinc finger protein 423 |
| chr3_+_141043050 | 0.78 |
ENST00000509842.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_-_149375783 | 0.77 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr10_+_5488564 | 0.77 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr1_+_100818009 | 0.76 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr19_+_41117770 | 0.76 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr11_-_76381029 | 0.75 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chrX_-_119695279 | 0.75 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr12_+_29376592 | 0.73 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr14_+_65878565 | 0.73 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr3_+_169940153 | 0.71 |
ENST00000295797.4
|
PRKCI
|
protein kinase C, iota |
| chr12_+_3068466 | 0.70 |
ENST00000358409.2
|
TEAD4
|
TEA domain family member 4 |
| chr4_+_129732467 | 0.70 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr1_+_100316041 | 0.70 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr2_-_61697862 | 0.70 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_+_29376673 | 0.69 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr18_-_53089723 | 0.69 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr20_+_35201993 | 0.69 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr12_+_3068544 | 0.69 |
ENST00000540314.1
ENST00000536826.1 ENST00000359864.2 |
TEAD4
|
TEA domain family member 4 |
| chr11_-_47470703 | 0.69 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr17_-_79105734 | 0.69 |
ENST00000417379.1
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr8_-_91657740 | 0.66 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr11_-_47470682 | 0.66 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr4_-_83719983 | 0.65 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr4_+_170581213 | 0.65 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr6_+_35996859 | 0.64 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr6_+_52930237 | 0.64 |
ENST00000323557.7
|
FBXO9
|
F-box protein 9 |
| chr1_-_94079648 | 0.64 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr14_+_24701628 | 0.63 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr5_-_38595498 | 0.63 |
ENST00000263409.4
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr17_-_67057203 | 0.63 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr16_+_2521500 | 0.63 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr11_-_47470591 | 0.63 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr6_+_123110302 | 0.62 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr18_-_53303123 | 0.62 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr15_-_52483566 | 0.61 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr22_-_37584321 | 0.61 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chrX_-_74376108 | 0.60 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr5_+_150157860 | 0.60 |
ENST00000600109.1
|
AC010441.1
|
AC010441.1 |
| chr1_+_154474689 | 0.60 |
ENST00000368482.4
|
TDRD10
|
tudor domain containing 10 |
| chr5_-_137071756 | 0.60 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr6_-_75915757 | 0.60 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr5_+_42423872 | 0.60 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr1_+_100818156 | 0.59 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr6_+_123110465 | 0.59 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr9_-_20622478 | 0.58 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr20_+_35201857 | 0.58 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr1_+_66458072 | 0.58 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_-_27630102 | 0.58 |
ENST00000356537.4
ENST00000522915.1 ENST00000539095.1 |
CCDC25
|
coiled-coil domain containing 25 |
| chr4_+_90800656 | 0.57 |
ENST00000394980.1
|
MMRN1
|
multimerin 1 |
| chr9_+_18474204 | 0.57 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr11_+_92085707 | 0.56 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr12_-_120687948 | 0.56 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr17_+_75181292 | 0.56 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr3_+_25469724 | 0.56 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr1_+_196621002 | 0.55 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr9_-_139891165 | 0.55 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr2_+_219125714 | 0.55 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr12_+_52445191 | 0.55 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_-_183387064 | 0.55 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_185538849 | 0.55 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr9_+_35749203 | 0.54 |
ENST00000456972.2
ENST00000378078.4 |
RGP1
|
RGP1 retrograde golgi transport homolog (S. cerevisiae) |
| chr10_-_17659234 | 0.54 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr14_-_23451467 | 0.54 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr14_-_77495007 | 0.53 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr8_-_141728760 | 0.53 |
ENST00000430260.2
|
PTK2
|
protein tyrosine kinase 2 |
| chr11_+_65190245 | 0.53 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr2_+_210444748 | 0.53 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_+_37856299 | 0.52 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr4_-_54457783 | 0.52 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr17_-_21156578 | 0.51 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr15_+_41549105 | 0.51 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr7_+_102715315 | 0.51 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr19_+_50691437 | 0.51 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chrX_-_33229636 | 0.50 |
ENST00000357033.4
|
DMD
|
dystrophin |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.1 | 3.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 1.0 | 7.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 4.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.7 | 2.2 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.7 | 2.0 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.6 | 4.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.6 | 2.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 5.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 2.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.5 | 1.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.5 | 16.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 1.3 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.4 | 1.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.4 | 1.1 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 | 1.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.3 | 1.0 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 3.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 1.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.3 | 0.9 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 0.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 3.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 0.9 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.3 | 4.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 1.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 1.6 | GO:0060214 | stem cell fate commitment(GO:0048865) endocardium formation(GO:0060214) |
| 0.2 | 1.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 0.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 1.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.2 | 1.9 | GO:0045602 | positive regulation of transcription of Notch receptor target(GO:0007221) negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 0.6 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.2 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.9 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 0.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 1.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 | 2.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 3.4 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.2 | 0.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 1.7 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.5 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 0.5 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 0.5 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 3.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.9 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 0.8 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 2.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 1.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.8 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.5 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 4.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 1.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.5 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.7 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 4.3 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.8 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.1 | 0.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.7 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 1.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 3.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.7 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.6 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.6 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0010949 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.3 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 1.0 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 8.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.7 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.0 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 3.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.0 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 1.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 3.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.8 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0003219 | secondary heart field specification(GO:0003139) cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0043586 | tongue development(GO:0043586) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.7 | 2.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.6 | 1.8 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.4 | 1.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 7.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 1.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 0.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 0.8 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 0.6 | GO:0097124 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 2.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 2.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.6 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 3.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 3.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 4.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 15.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 5.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 1.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 10.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 2.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.8 | 2.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.7 | 2.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.6 | 16.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 12.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 3.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 1.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 3.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.7 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 1.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 1.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.7 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.4 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 4.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.7 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 1.6 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 1.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.6 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 5.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 2.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 2.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 3.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 13.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 2.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 6.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0008430 | thyroxine 5'-deiodinase activity(GO:0004800) selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 15.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 12.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 7.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 6.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 7.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.5 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 1.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 3.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 5.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |