Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
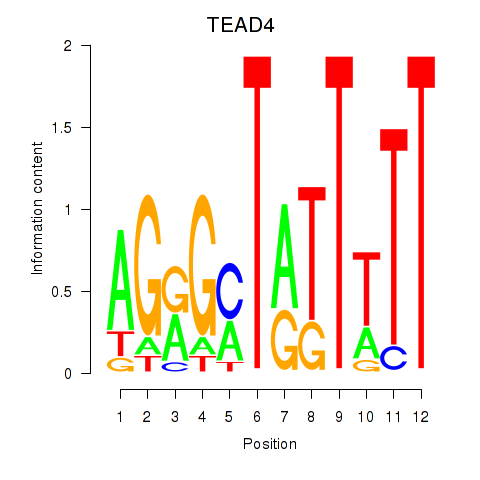
Results for TEAD4
Z-value: 0.85
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3068544_3068597, hg19_v2_chr12_+_3069037_3069119, hg19_v2_chr12_+_3068466_3068496 | 0.73 | 2.9e-05 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41614909 | 4.32 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41614720 | 3.84 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_18466026 | 2.92 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr4_-_159080806 | 2.63 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr4_-_186732048 | 2.56 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_-_69113218 | 2.45 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr2_+_38893208 | 2.29 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr2_-_136875712 | 2.17 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr5_-_111093167 | 2.09 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr9_+_2029019 | 2.04 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_61547894 | 2.01 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr9_+_71944241 | 1.99 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_79258547 | 1.98 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr4_-_110723134 | 1.94 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr1_+_164528866 | 1.93 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr11_-_118134997 | 1.92 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr4_-_89152474 | 1.88 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_+_90098606 | 1.82 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr4_-_149365827 | 1.82 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr12_+_79258444 | 1.79 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr21_+_17443434 | 1.61 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_110723335 | 1.58 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_110723194 | 1.51 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr21_+_17909594 | 1.51 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_9913489 | 1.43 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr21_+_17443521 | 1.33 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_66951474 | 1.32 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_-_70994612 | 1.28 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_+_13141097 | 1.25 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr18_+_3466248 | 1.17 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr17_+_47865917 | 1.15 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr2_+_238767517 | 1.11 |
ENST00000404910.2
|
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1 |
| chr18_+_6834472 | 1.06 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr14_-_89883412 | 1.03 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr9_+_90112117 | 1.01 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_+_66458072 | 0.97 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_-_112356944 | 0.94 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr21_-_27423339 | 0.90 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr1_-_154928562 | 0.90 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr11_-_63376013 | 0.89 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr2_+_191792376 | 0.88 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr14_+_76071805 | 0.87 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr7_-_37024665 | 0.84 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_+_41698927 | 0.83 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr21_+_47706537 | 0.81 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr3_+_123813509 | 0.79 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr12_+_48876275 | 0.77 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr3_+_132316081 | 0.75 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr2_+_38893047 | 0.75 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr2_+_1418154 | 0.67 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr2_-_188419200 | 0.67 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr13_+_78315466 | 0.65 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_-_13328564 | 0.61 |
ENST00000606530.1
ENST00000607658.1 ENST00000343141.4 ENST00000356436.4 ENST00000379300.3 ENST00000452989.1 ENST00000450347.1 ENST00000422136.1 ENST00000446018.1 ENST00000379291.1 ENST00000379307.2 ENST00000606370.1 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr20_-_23402028 | 0.61 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr1_-_227505289 | 0.61 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr21_-_39870339 | 0.60 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr9_+_75229616 | 0.60 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr11_+_120255997 | 0.59 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr12_+_123011776 | 0.57 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr14_+_31046959 | 0.55 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr20_+_1246908 | 0.55 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr14_-_106791536 | 0.55 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr1_+_163038565 | 0.55 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr14_-_75530693 | 0.54 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chrX_+_37639264 | 0.54 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr12_-_58220078 | 0.52 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr6_+_96025341 | 0.52 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr2_-_188419078 | 0.51 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_+_241695670 | 0.51 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_-_10588595 | 0.49 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr15_-_20193370 | 0.47 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr1_+_241695424 | 0.46 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_99279928 | 0.46 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chrX_-_10588459 | 0.46 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_-_164875850 | 0.43 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr5_-_10308125 | 0.43 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr14_+_21236586 | 0.42 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr9_-_33402506 | 0.41 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr13_-_37679803 | 0.41 |
ENST00000379800.3
|
CSNK1A1L
|
casein kinase 1, alpha 1-like |
| chr17_+_38219063 | 0.41 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr6_-_159466042 | 0.40 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr12_+_32260085 | 0.40 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr2_+_238768187 | 0.40 |
ENST00000254661.4
ENST00000409726.1 |
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1 |
| chr3_-_185538849 | 0.39 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr8_+_107282368 | 0.39 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr11_+_17316870 | 0.39 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr7_+_80267973 | 0.38 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_+_20963632 | 0.37 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr11_+_57308979 | 0.37 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr12_+_20963647 | 0.37 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_+_122720681 | 0.37 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr1_+_206516200 | 0.35 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chrX_+_79675965 | 0.35 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr8_+_67405755 | 0.35 |
ENST00000521495.1
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr19_+_9296279 | 0.35 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr5_-_138775177 | 0.34 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_+_57878859 | 0.33 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr14_-_106518922 | 0.33 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr1_+_41204506 | 0.33 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr2_-_211341411 | 0.33 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr19_+_54369608 | 0.32 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chr14_-_37051798 | 0.32 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr12_-_91572278 | 0.32 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr12_-_91539918 | 0.32 |
ENST00000548218.1
|
DCN
|
decorin |
| chr3_+_101546827 | 0.31 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chrX_+_52240504 | 0.31 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr3_-_156878540 | 0.31 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr4_+_87928413 | 0.31 |
ENST00000544085.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr7_-_14029283 | 0.31 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr1_-_59249732 | 0.31 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr3_+_123813543 | 0.31 |
ENST00000360013.3
|
KALRN
|
kalirin, RhoGEF kinase |
| chr12_-_123011476 | 0.30 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr1_-_207119738 | 0.30 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr7_-_82073031 | 0.30 |
ENST00000356253.5
ENST00000423588.1 |
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr1_-_60392452 | 0.30 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr2_-_20251744 | 0.29 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr5_+_53751445 | 0.29 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr11_-_104916034 | 0.27 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr8_+_42128812 | 0.27 |
ENST00000520810.1
ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr17_+_44588877 | 0.27 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr14_-_45252031 | 0.25 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr4_+_141264597 | 0.25 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr5_-_54281491 | 0.25 |
ENST00000381405.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr7_+_77167343 | 0.24 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr4_-_144826682 | 0.24 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr6_-_52705641 | 0.24 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr2_-_196933536 | 0.23 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr5_+_118812294 | 0.23 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr19_-_41256207 | 0.23 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr19_-_50083803 | 0.23 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chrX_-_52258669 | 0.23 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chrX_+_37639302 | 0.22 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr19_-_50083822 | 0.22 |
ENST00000596358.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chr3_+_197518100 | 0.22 |
ENST00000438796.2
ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr10_-_50970322 | 0.22 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chrX_+_22056165 | 0.22 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chrX_+_21392553 | 0.21 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr16_+_4364762 | 0.21 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr10_-_50970382 | 0.21 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr13_-_62001982 | 0.20 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr4_-_186317034 | 0.20 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr6_-_29399744 | 0.20 |
ENST00000377154.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr11_-_6440624 | 0.19 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr14_-_106622419 | 0.19 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr10_+_63808970 | 0.19 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chrX_+_21392529 | 0.19 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr7_+_37960163 | 0.19 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr4_+_71588372 | 0.18 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_-_19144243 | 0.18 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr7_+_23338819 | 0.18 |
ENST00000466681.1
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr7_+_150811705 | 0.17 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr2_-_235405679 | 0.17 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr19_+_37825526 | 0.16 |
ENST00000592768.1
ENST00000591417.1 ENST00000585623.1 ENST00000592168.1 ENST00000591391.1 ENST00000392153.3 ENST00000589392.1 ENST00000324411.4 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr1_-_87379785 | 0.16 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr2_+_108905325 | 0.16 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_+_130279178 | 0.15 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr1_-_247697141 | 0.15 |
ENST00000366487.3
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3 |
| chr16_-_20681177 | 0.15 |
ENST00000524149.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr10_-_75410771 | 0.15 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr10_+_47894572 | 0.15 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr19_+_17403413 | 0.15 |
ENST00000595444.1
ENST00000600434.1 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr4_+_141178440 | 0.14 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr1_+_109256067 | 0.14 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr19_+_17862274 | 0.14 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr12_-_123011536 | 0.14 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr2_-_161056762 | 0.13 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr18_-_67873078 | 0.13 |
ENST00000255674.6
|
RTTN
|
rotatin |
| chr9_-_104249400 | 0.13 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr14_-_106845789 | 0.13 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr11_+_12766583 | 0.12 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr13_+_78315295 | 0.12 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_37837185 | 0.12 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr1_+_46016703 | 0.12 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr4_-_26492076 | 0.11 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr6_-_41039567 | 0.11 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr12_-_54652060 | 0.11 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr10_+_5005445 | 0.11 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr13_+_113760098 | 0.11 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr20_+_33814457 | 0.11 |
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted) |
| chr4_-_155533787 | 0.11 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr18_-_67872891 | 0.11 |
ENST00000454359.1
ENST00000437017.1 |
RTTN
|
rotatin |
| chr1_+_164529004 | 0.11 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_+_120629653 | 0.11 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_123488124 | 0.10 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr5_+_138609782 | 0.10 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr11_+_125496400 | 0.10 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr1_-_226187013 | 0.10 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr6_-_42690312 | 0.10 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr9_+_90497741 | 0.10 |
ENST00000325643.5
|
SPATA31E1
|
SPATA31 subfamily E, member 1 |
| chrX_+_21392873 | 0.10 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr21_+_47878757 | 0.10 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr3_+_38307293 | 0.10 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr17_+_17685422 | 0.10 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr18_+_61254570 | 0.09 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr6_-_32634425 | 0.09 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr4_+_70861647 | 0.09 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr15_-_52263937 | 0.09 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr7_-_32529973 | 0.08 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_158656488 | 0.08 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr7_-_14028488 | 0.08 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.8 | 3.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 1.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.4 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 2.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 3.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.6 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 1.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 2.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 0.8 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 1.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0045925 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.4 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 | 0.6 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 2.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 2.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 2.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.9 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 1.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 2.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 5.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 4.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 2.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.5 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.4 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 2.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 1.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.4 | 3.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 0.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 2.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 0.9 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 3.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 3.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 5.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.8 | GO:0050664 | superoxide-generating NADPH oxidase activity(GO:0016175) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 3.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 5.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |