Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for TFEC_MITF_ARNTL_BHLHE41
Z-value: 0.90
Transcription factors associated with TFEC_MITF_ARNTL_BHLHE41
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
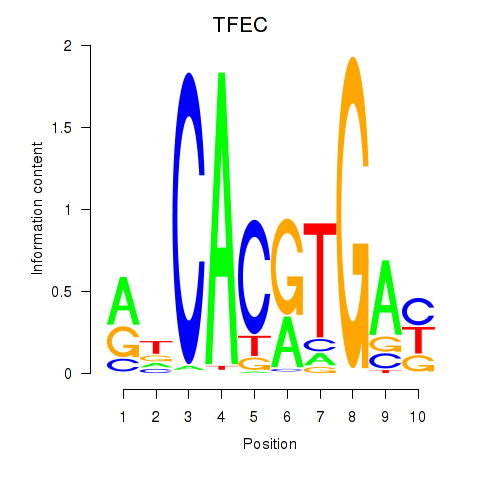
TFEC
|
ENSG00000105967.11 | transcription factor EC |
|
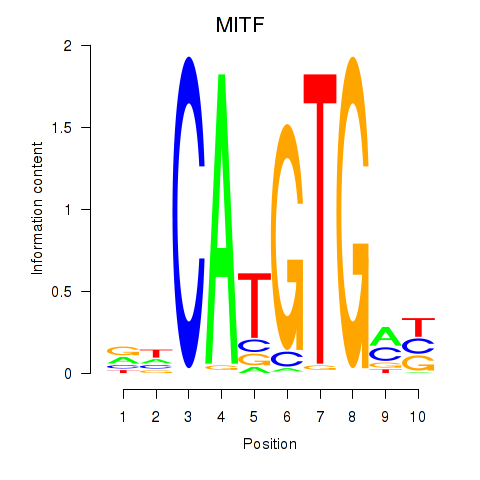
MITF
|
ENSG00000187098.10 | melanocyte inducing transcription factor |
|
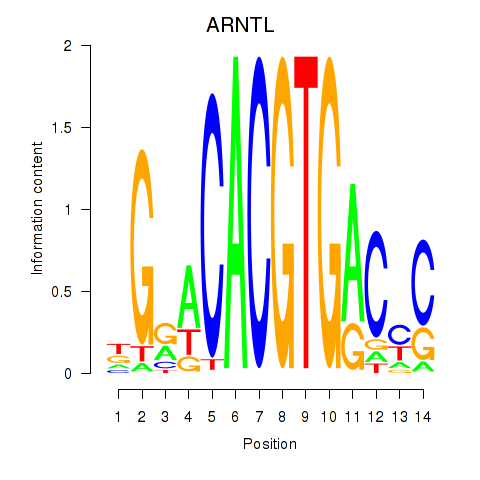
ARNTL
|
ENSG00000133794.13 | aryl hydrocarbon receptor nuclear translocator like |
|
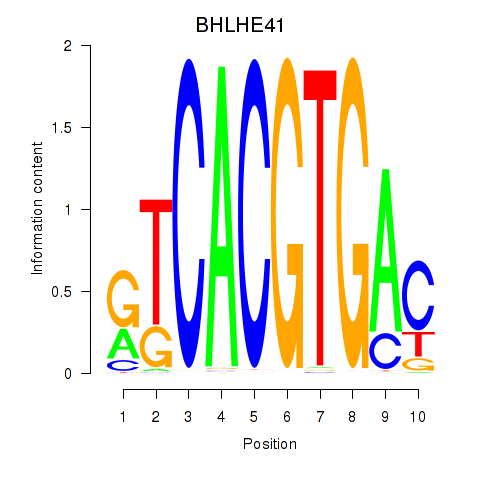
BHLHE41
|
ENSG00000123095.5 | basic helix-loop-helix family member e41 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MITF | hg19_v2_chr3_+_69812877_69812962, hg19_v2_chr3_+_69928256_69928385 | -0.37 | 6.8e-02 | Click! |
| BHLHE41 | hg19_v2_chr12_-_26278030_26278060 | -0.10 | 6.5e-01 | Click! |
| TFEC | hg19_v2_chr7_-_115670804_115670867, hg19_v2_chr7_-_115608304_115608352 | 0.07 | 7.3e-01 | Click! |
| ARNTL | hg19_v2_chr11_+_13299186_13299432 | 0.06 | 7.9e-01 | Click! |
Activity profile of TFEC_MITF_ARNTL_BHLHE41 motif
Sorted Z-values of TFEC_MITF_ARNTL_BHLHE41 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFEC_MITF_ARNTL_BHLHE41
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 3.4 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.7 | 2.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.4 | 2.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 1.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.3 | 0.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.3 | 0.9 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 2.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 1.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.6 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 1.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.6 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 0.6 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.6 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.7 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.2 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.2 | 2.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.4 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 1.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 2.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.6 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.8 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 1.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.3 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.3 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.4 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.1 | 3.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.3 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 1.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 0.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.3 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 1.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.5 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 0.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.5 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 2.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.4 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 1.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) cellular response to camptothecin(GO:0072757) |
| 0.1 | 1.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 0.5 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 2.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 1.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.3 | GO:0034033 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 2.5 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.0 | 0.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 1.3 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.9 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.0 | 0.4 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.7 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.0 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0003214 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.6 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:1900195 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:1902903 | regulation of fibril organization(GO:1902903) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.3 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.3 | 0.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 2.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 2.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 2.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 2.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 3.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.5 | 1.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 2.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.6 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.2 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 0.6 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 3.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.6 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 1.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0031177 | S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 2.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |