Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for VAX2_RHOXF2
Z-value: 0.40
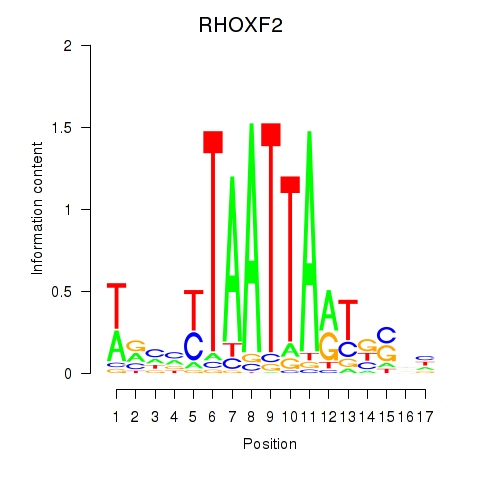
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX2
|
ENSG00000116035.2 | ventral anterior homeobox 2 |
|
RHOXF2
|
ENSG00000131721.4 | Rhox homeobox family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX2 | hg19_v2_chr2_+_71127699_71127744 | 0.30 | 1.5e-01 | Click! |
| RHOXF2 | hg19_v2_chrX_+_119292467_119292467 | -0.29 | 1.7e-01 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186696425 | 1.68 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_35258592 | 0.94 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr6_+_130339710 | 0.51 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr12_-_10022735 | 0.41 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr3_-_112127981 | 0.41 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr20_-_29978286 | 0.36 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr3_-_141747950 | 0.34 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr9_-_3469181 | 0.34 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr17_-_41466555 | 0.33 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr19_-_36001113 | 0.31 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr4_-_138453606 | 0.28 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr4_+_66536248 | 0.27 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr7_+_12727250 | 0.26 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr2_+_103089756 | 0.24 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr20_-_33735070 | 0.23 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_+_62439037 | 0.22 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_211307404 | 0.22 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr12_+_131438443 | 0.22 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr1_-_190446759 | 0.20 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr2_-_198540719 | 0.19 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr11_+_71900703 | 0.19 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_71900572 | 0.19 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr2_-_74618964 | 0.19 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr16_-_30122717 | 0.19 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_-_74619152 | 0.19 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr11_-_5323226 | 0.17 |
ENST00000380224.1
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4 |
| chr13_+_36050881 | 0.16 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr10_+_6779326 | 0.16 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr17_+_43238438 | 0.15 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr1_-_201140673 | 0.15 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chrX_-_18690210 | 0.14 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr9_-_95166841 | 0.14 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr9_+_125133315 | 0.14 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_125132803 | 0.14 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr10_-_50396407 | 0.13 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr3_-_151034734 | 0.13 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr15_+_74218787 | 0.13 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr4_-_66536196 | 0.12 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr1_-_67266939 | 0.12 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr20_-_50418947 | 0.12 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_49462434 | 0.11 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr20_-_50418972 | 0.11 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr5_+_176811431 | 0.11 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr15_+_49462397 | 0.11 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr7_+_99717230 | 0.10 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr4_+_88754113 | 0.10 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr9_-_21202204 | 0.10 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr17_-_38545799 | 0.10 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr14_+_32798547 | 0.10 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_-_138453559 | 0.10 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_80351910 | 0.09 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr15_+_80351977 | 0.09 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_-_17171817 | 0.09 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr17_-_39203519 | 0.09 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr12_-_9760482 | 0.09 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr16_+_53133070 | 0.09 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chrX_+_72783026 | 0.09 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr15_+_58702742 | 0.09 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr21_+_17443434 | 0.09 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_48823896 | 0.08 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr15_-_89755034 | 0.08 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_+_153747746 | 0.08 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr4_+_159727272 | 0.08 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr7_+_115862858 | 0.08 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr21_+_17443521 | 0.08 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_76481258 | 0.08 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr20_-_50419055 | 0.07 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr14_-_39639523 | 0.07 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr14_+_74034310 | 0.07 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr1_-_24151903 | 0.07 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr21_+_17442799 | 0.07 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_+_11071546 | 0.07 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr6_+_3259148 | 0.07 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chrX_+_135730297 | 0.07 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr4_-_25865159 | 0.06 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr9_-_5304432 | 0.06 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chrX_+_43515467 | 0.06 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr5_+_129083772 | 0.06 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr9_-_95166884 | 0.06 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr12_-_10959892 | 0.06 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr5_-_89770582 | 0.06 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr1_+_54569968 | 0.06 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr5_+_174151536 | 0.06 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr9_+_12693336 | 0.06 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chrX_+_135730373 | 0.06 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr17_-_38821373 | 0.06 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr7_-_44122063 | 0.06 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr5_+_89770664 | 0.05 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chrX_+_7137475 | 0.05 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr17_-_18430160 | 0.05 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr4_+_80584903 | 0.05 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr5_+_89770696 | 0.05 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr12_-_6233828 | 0.05 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr18_-_44181442 | 0.05 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr19_+_50016610 | 0.05 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_191000172 | 0.05 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr1_-_212965104 | 0.05 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr20_-_29978383 | 0.04 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_212965170 | 0.04 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr17_+_48823975 | 0.04 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr4_-_159956333 | 0.04 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr11_-_111794446 | 0.04 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr17_+_40610862 | 0.04 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr9_-_21239978 | 0.03 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr6_-_82957433 | 0.03 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr4_-_74486347 | 0.03 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr17_-_10372875 | 0.03 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr20_+_15177480 | 0.03 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr2_-_85555385 | 0.03 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr9_+_131062367 | 0.03 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr4_-_74486109 | 0.03 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_99717463 | 0.03 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_+_158519654 | 0.03 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr7_+_138145076 | 0.02 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr1_-_152386732 | 0.02 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr12_+_31812121 | 0.02 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr4_-_69111401 | 0.02 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr16_-_28608424 | 0.02 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr2_+_228736321 | 0.02 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr7_+_148936732 | 0.02 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr14_+_56584414 | 0.02 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr3_+_157154578 | 0.02 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr10_-_99205607 | 0.02 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr7_+_100860949 | 0.02 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr4_-_66536057 | 0.01 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr11_+_101918153 | 0.01 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr4_+_88896819 | 0.01 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr7_+_73245193 | 0.01 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr8_-_7243080 | 0.01 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr7_-_121944491 | 0.01 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr15_+_58430567 | 0.01 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr12_+_16500037 | 0.01 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr7_-_100860851 | 0.01 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr12_-_50643664 | 0.01 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr15_+_58430368 | 0.01 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr14_+_20923350 | 0.01 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr2_-_40680578 | 0.01 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_-_149829244 | 0.01 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr16_+_72088376 | 0.01 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr5_-_20575959 | 0.00 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr5_-_149829294 | 0.00 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr7_-_111032971 | 0.00 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr5_-_149829314 | 0.00 |
ENST00000407193.1
|
RPS14
|
ribosomal protein S14 |
| chr12_+_81110684 | 0.00 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr3_+_152552685 | 0.00 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 1.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.9 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |