Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
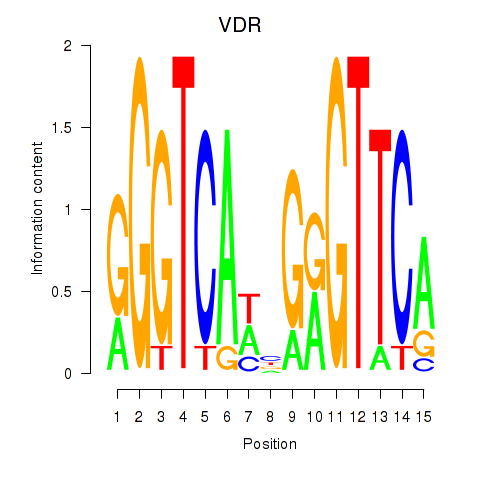
Results for VDR
Z-value: 0.62
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | vitamin D receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | 0.03 | 8.8e-01 | Click! |
Activity profile of VDR motif
Sorted Z-values of VDR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_53313905 | 0.86 |
ENST00000377962.3
ENST00000448904.2 |
LECT1
|
leukocyte cell derived chemotaxin 1 |
| chr3_+_54157480 | 0.66 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr17_-_34807272 | 0.52 |
ENST00000535592.1
ENST00000394453.1 |
TBC1D3G
|
TBC1 domain family, member 3G |
| chr17_-_34808047 | 0.52 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr17_-_36347835 | 0.50 |
ENST00000519532.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_-_34756219 | 0.49 |
ENST00000451448.2
ENST00000394359.3 |
TBC1D3C
TBC1D3H
|
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr17_-_36348610 | 0.49 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr8_-_93115445 | 0.48 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_13261216 | 0.47 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr17_+_36284791 | 0.46 |
ENST00000505415.1
|
TBC1D3F
|
TBC1 domain family, member 3F |
| chr16_-_67260691 | 0.46 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr11_+_65266507 | 0.45 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr14_-_35873856 | 0.43 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr12_-_25348007 | 0.42 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr1_+_61869748 | 0.39 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr16_-_75282088 | 0.35 |
ENST00000542031.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr21_+_25801041 | 0.34 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr16_-_30107491 | 0.32 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr17_-_34591208 | 0.31 |
ENST00000336331.5
|
TBC1D3C
|
TBC1 domain family, member 3C |
| chr6_-_32145101 | 0.31 |
ENST00000375104.2
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr22_+_18593507 | 0.30 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr6_-_42016385 | 0.30 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr2_+_230787213 | 0.29 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr2_+_230787201 | 0.28 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr2_-_25194476 | 0.28 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr10_-_18940501 | 0.27 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr16_-_67281413 | 0.27 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr19_+_49122785 | 0.26 |
ENST00000598088.1
|
SPHK2
|
sphingosine kinase 2 |
| chr1_+_35258592 | 0.26 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr6_+_125475335 | 0.26 |
ENST00000532429.1
ENST00000534199.1 |
TPD52L1
|
tumor protein D52-like 1 |
| chr12_+_3069037 | 0.26 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr6_+_35227449 | 0.26 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr13_+_24844819 | 0.25 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr1_+_200993071 | 0.25 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr11_-_73882029 | 0.25 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr1_-_205091115 | 0.25 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr12_-_56848426 | 0.25 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr1_+_44457261 | 0.24 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24 |
| chr7_-_36764004 | 0.24 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr17_+_56270084 | 0.23 |
ENST00000225371.5
|
EPX
|
eosinophil peroxidase |
| chr9_+_131644781 | 0.23 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr16_-_68404908 | 0.23 |
ENST00000574662.1
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr19_+_49122548 | 0.23 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr13_+_57721622 | 0.22 |
ENST00000377930.1
|
PRR20B
|
proline rich 20B |
| chrX_+_153770421 | 0.22 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr7_+_155089486 | 0.22 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr16_+_14802801 | 0.21 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr11_+_60869867 | 0.21 |
ENST00000347785.3
|
CD5
|
CD5 molecule |
| chr11_+_124609823 | 0.21 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr17_+_68100989 | 0.20 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_25874440 | 0.20 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr7_-_105029812 | 0.20 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr7_-_36764142 | 0.20 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr16_-_2770216 | 0.19 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr12_-_58135903 | 0.19 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr9_+_71789133 | 0.19 |
ENST00000348208.4
ENST00000265384.7 |
TJP2
|
tight junction protein 2 |
| chr16_-_67217844 | 0.19 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr15_+_40226304 | 0.19 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr19_-_54693401 | 0.19 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_+_133861790 | 0.18 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr7_-_16844611 | 0.18 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr15_-_31283798 | 0.18 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr6_+_30525051 | 0.18 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr5_-_177210399 | 0.17 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr16_+_1128781 | 0.17 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr10_-_33623826 | 0.17 |
ENST00000374867.2
|
NRP1
|
neuropilin 1 |
| chr15_-_59225844 | 0.17 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr6_+_131521120 | 0.17 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_177134134 | 0.17 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr17_-_43487741 | 0.17 |
ENST00000455881.1
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr11_-_61582579 | 0.17 |
ENST00000539419.1
ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1
|
fatty acid desaturase 1 |
| chr17_-_7082861 | 0.17 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_-_506316 | 0.16 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr3_+_48264816 | 0.16 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr14_+_100531615 | 0.16 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr19_+_18530146 | 0.16 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr5_+_175487692 | 0.16 |
ENST00000510151.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr11_-_4629388 | 0.16 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr6_-_117086873 | 0.16 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr15_+_41851211 | 0.16 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr6_+_30524663 | 0.16 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr22_-_29663954 | 0.16 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr19_-_47987419 | 0.16 |
ENST00000536339.1
ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN
|
kaptin (actin binding protein) |
| chr14_+_95047744 | 0.16 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr8_+_24298597 | 0.15 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr5_+_177433973 | 0.15 |
ENST00000507848.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr17_-_6616678 | 0.15 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr10_+_54074033 | 0.15 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_+_24298531 | 0.15 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr1_-_19426149 | 0.15 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr10_-_106240032 | 0.15 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr14_+_89290965 | 0.15 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr17_+_27895609 | 0.15 |
ENST00000581411.2
ENST00000301057.7 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr15_-_72563585 | 0.14 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_-_72722783 | 0.14 |
ENST00000428206.1
ENST00000252594.6 ENST00000310326.8 ENST00000438747.2 |
NSUN5
|
NOP2/Sun domain family, member 5 |
| chr19_-_11494975 | 0.14 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr14_-_91884115 | 0.14 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr22_-_24126145 | 0.14 |
ENST00000598975.1
|
AP000349.1
|
Uncharacterized protein |
| chr5_-_179051579 | 0.14 |
ENST00000505811.1
ENST00000515714.1 ENST00000513225.1 ENST00000503664.1 ENST00000356731.5 ENST00000523137.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr1_-_205326022 | 0.14 |
ENST00000367155.3
|
KLHDC8A
|
kelch domain containing 8A |
| chr16_+_31119615 | 0.14 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr12_+_52695617 | 0.14 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr17_-_43487780 | 0.14 |
ENST00000532038.1
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr19_+_7599597 | 0.14 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr11_+_64808675 | 0.14 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr9_-_34376851 | 0.14 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr20_+_30028322 | 0.14 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr3_-_178969403 | 0.14 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr4_-_36246060 | 0.13 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr16_+_532503 | 0.13 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr1_-_205326161 | 0.13 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr20_+_58630972 | 0.13 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr15_-_59225758 | 0.13 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chr2_-_203736452 | 0.13 |
ENST00000419460.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr10_-_104001231 | 0.13 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr11_-_111794446 | 0.12 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr7_+_155090271 | 0.12 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr2_+_210517895 | 0.12 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr19_-_5719860 | 0.12 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr11_+_73882311 | 0.12 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr20_+_48909240 | 0.12 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr3_+_13521665 | 0.12 |
ENST00000295757.3
ENST00000402259.1 ENST00000402271.1 ENST00000446613.2 ENST00000404548.1 ENST00000404040.1 |
HDAC11
|
histone deacetylase 11 |
| chr10_-_5541525 | 0.12 |
ENST00000380332.3
|
CALML5
|
calmodulin-like 5 |
| chr9_+_131038425 | 0.11 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr8_-_13134045 | 0.11 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr8_-_135522425 | 0.11 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr4_-_11430221 | 0.11 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr9_+_111624577 | 0.11 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr12_-_114404111 | 0.11 |
ENST00000545145.2
ENST00000392561.3 ENST00000261741.5 |
RBM19
|
RNA binding motif protein 19 |
| chr3_-_27763803 | 0.11 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr8_+_119294456 | 0.11 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr11_-_65667997 | 0.11 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr12_-_96390063 | 0.11 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr11_+_73882144 | 0.11 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr12_+_57857475 | 0.11 |
ENST00000528467.1
|
GLI1
|
GLI family zinc finger 1 |
| chr22_+_39077947 | 0.11 |
ENST00000216034.4
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr22_+_23213658 | 0.11 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr7_-_151433393 | 0.11 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr2_+_38893208 | 0.11 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr7_-_151433342 | 0.11 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr12_+_110906169 | 0.11 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr19_+_39936267 | 0.11 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr3_-_27764190 | 0.11 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr5_-_177209832 | 0.11 |
ENST00000393518.3
ENST00000505531.1 ENST00000503567.1 |
FAM153A
|
family with sequence similarity 153, member A |
| chr2_+_233527443 | 0.10 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr11_-_65667884 | 0.10 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr11_-_6624801 | 0.10 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr11_-_82782861 | 0.10 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr19_+_39936186 | 0.10 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr20_-_23549334 | 0.10 |
ENST00000376979.3
|
CST9L
|
cystatin 9-like |
| chr5_+_175488258 | 0.10 |
ENST00000512862.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr9_-_139839064 | 0.10 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr2_-_25194963 | 0.10 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr2_-_85625857 | 0.10 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr19_+_41305085 | 0.10 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_-_87799505 | 0.10 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr14_-_106781017 | 0.10 |
ENST00000390612.2
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr10_-_33623564 | 0.10 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr4_+_144434584 | 0.10 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr15_+_69307028 | 0.10 |
ENST00000388866.3
ENST00000530406.2 |
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr9_+_130213774 | 0.09 |
ENST00000373324.4
ENST00000323301.4 |
LRSAM1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr10_+_47894572 | 0.09 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr7_+_99699280 | 0.09 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr19_+_14017116 | 0.09 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr19_+_50270219 | 0.09 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr8_+_23104130 | 0.09 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr19_+_41281060 | 0.09 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr2_+_220363579 | 0.09 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr12_+_48876275 | 0.09 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr1_+_26348259 | 0.09 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr1_+_153232160 | 0.09 |
ENST00000368742.3
|
LOR
|
loricrin |
| chr8_+_75736761 | 0.09 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr20_+_42193856 | 0.09 |
ENST00000412111.1
ENST00000423407.3 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr13_-_38172863 | 0.09 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr5_-_179227540 | 0.09 |
ENST00000520875.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr12_-_96390108 | 0.09 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr16_+_28834303 | 0.08 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr1_+_100111580 | 0.08 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr9_-_139343294 | 0.08 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr3_-_93781750 | 0.08 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr11_+_28129795 | 0.08 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr3_+_157154578 | 0.08 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr16_-_28506840 | 0.08 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr1_+_92414952 | 0.08 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr1_+_109234907 | 0.08 |
ENST00000370025.4
ENST00000370022.5 ENST00000370021.1 |
PRPF38B
|
pre-mRNA processing factor 38B |
| chr19_-_49122384 | 0.08 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr19_+_41281282 | 0.08 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr2_-_232329186 | 0.08 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr1_+_36348790 | 0.08 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr10_-_22292675 | 0.08 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr16_+_88636789 | 0.08 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr14_+_105212297 | 0.08 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr12_+_53895364 | 0.08 |
ENST00000552817.1
ENST00000394357.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr10_-_76995769 | 0.08 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr13_+_113656022 | 0.07 |
ENST00000423482.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr2_+_176957619 | 0.07 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chrX_+_149737046 | 0.07 |
ENST00000370396.2
ENST00000542741.1 ENST00000543350.1 ENST00000424519.1 ENST00000413012.2 |
MTM1
|
myotubularin 1 |
| chr2_+_38893047 | 0.07 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr16_-_28937027 | 0.07 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr15_+_75940218 | 0.07 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.2 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.2 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 3.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.2 | GO:0090154 | sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1990523 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.4 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |