Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
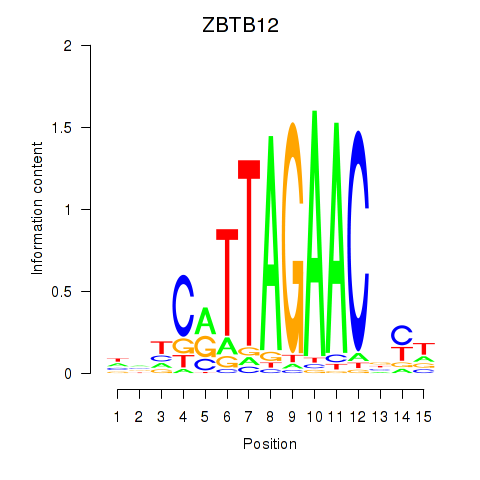
Results for ZBTB12
Z-value: 0.54
Transcription factors associated with ZBTB12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB12
|
ENSG00000204366.3 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB12 | hg19_v2_chr6_-_31869769_31869880 | -0.30 | 1.4e-01 | Click! |
Activity profile of ZBTB12 motif
Sorted Z-values of ZBTB12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_61869748 | 2.08 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr14_-_91526462 | 2.04 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr1_+_158979686 | 0.79 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979680 | 0.78 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979792 | 0.78 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_-_88179302 | 0.78 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_+_147527103 | 0.75 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr3_-_149051444 | 0.73 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr6_+_18387570 | 0.68 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr5_-_88178964 | 0.65 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr1_+_84873913 | 0.63 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease II beta |
| chr12_-_46766577 | 0.62 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr5_-_88179017 | 0.60 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr3_-_164875850 | 0.59 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr3_-_149051194 | 0.59 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr14_-_21270995 | 0.58 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr9_-_80437915 | 0.56 |
ENST00000397476.3
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr1_+_32757668 | 0.53 |
ENST00000373548.3
|
HDAC1
|
histone deacetylase 1 |
| chr2_+_190722119 | 0.53 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr7_-_93520191 | 0.51 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr19_+_55417499 | 0.51 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr2_+_89999259 | 0.49 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr8_+_118533049 | 0.48 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr8_+_1993152 | 0.47 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr17_-_39677971 | 0.47 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr10_+_5454505 | 0.47 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr17_-_5487768 | 0.45 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr8_+_118532937 | 0.45 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chrX_-_133931164 | 0.44 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chrX_-_73072534 | 0.43 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr1_+_46049706 | 0.39 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr17_+_4643337 | 0.38 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr20_-_36793663 | 0.38 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr9_+_127615733 | 0.37 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chrX_+_70338525 | 0.36 |
ENST00000374102.1
|
MED12
|
mediator complex subunit 12 |
| chr8_+_1993173 | 0.36 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr19_+_55417530 | 0.34 |
ENST00000350790.5
ENST00000338835.5 ENST00000357397.5 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr6_+_2988847 | 0.34 |
ENST00000380472.3
ENST00000605901.1 ENST00000454015.1 |
NQO2
LINC01011
|
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr11_-_2170786 | 0.30 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr7_-_93520259 | 0.30 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr5_-_38557561 | 0.29 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr13_-_37633743 | 0.29 |
ENST00000497318.1
ENST00000475892.1 ENST00000356185.3 ENST00000350612.6 ENST00000542180.1 ENST00000360252.4 |
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr18_-_53177984 | 0.29 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr11_+_63449045 | 0.28 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr3_-_114343039 | 0.25 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr5_-_148929848 | 0.25 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr1_+_201755452 | 0.24 |
ENST00000438083.1
|
NAV1
|
neuron navigator 1 |
| chr12_-_11214893 | 0.24 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr15_-_34880646 | 0.24 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr19_-_11688500 | 0.23 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr11_+_112047087 | 0.22 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr17_-_46178527 | 0.22 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr18_-_24445664 | 0.21 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr20_-_25320367 | 0.21 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chrX_-_24690771 | 0.20 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr14_-_50154921 | 0.20 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr19_-_46148820 | 0.20 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr12_-_58329819 | 0.19 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr2_+_173600514 | 0.19 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_54908043 | 0.18 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr11_-_59952106 | 0.17 |
ENST00000529054.1
ENST00000530839.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_-_170599561 | 0.17 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr11_+_12132117 | 0.16 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr14_-_70826438 | 0.16 |
ENST00000389912.6
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr10_+_23217006 | 0.15 |
ENST00000376528.4
ENST00000447081.1 |
ARMC3
|
armadillo repeat containing 3 |
| chr7_-_142139783 | 0.15 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr10_+_23216944 | 0.15 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chr14_-_22938665 | 0.15 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr12_-_58329888 | 0.14 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr3_+_101818088 | 0.14 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr5_-_122372354 | 0.14 |
ENST00000306442.4
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C) |
| chr19_+_46195895 | 0.14 |
ENST00000366382.4
|
QPCTL
|
glutaminyl-peptide cyclotransferase-like |
| chr22_-_42084863 | 0.13 |
ENST00000401959.1
ENST00000355257.3 |
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr12_+_53689309 | 0.13 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr17_-_61781750 | 0.13 |
ENST00000582026.1
|
STRADA
|
STE20-related kinase adaptor alpha |
| chr12_+_123459127 | 0.13 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr2_+_234545092 | 0.13 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_-_109203997 | 0.12 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr11_-_101000445 | 0.11 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr6_+_36097992 | 0.11 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr19_-_51531210 | 0.11 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr1_-_198990166 | 0.10 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr11_+_60145948 | 0.10 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr20_-_33543546 | 0.10 |
ENST00000216951.2
|
GSS
|
glutathione synthetase |
| chr3_+_26735991 | 0.10 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr10_+_180405 | 0.09 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr1_+_22778337 | 0.09 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr7_-_138347897 | 0.09 |
ENST00000288513.5
|
SVOPL
|
SVOP-like |
| chr1_+_158901329 | 0.09 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr12_+_54378923 | 0.09 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr18_-_32870148 | 0.08 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr17_-_27405875 | 0.08 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr1_+_100598691 | 0.08 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr14_-_74959978 | 0.08 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr3_-_151047327 | 0.07 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr12_+_49121213 | 0.07 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr22_+_22930626 | 0.07 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr6_+_26158343 | 0.07 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr14_-_74959994 | 0.07 |
ENST00000238633.2
ENST00000434013.2 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr8_+_110098850 | 0.07 |
ENST00000518632.1
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr19_+_11485333 | 0.07 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr10_+_180643 | 0.07 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr5_+_148786423 | 0.07 |
ENST00000505254.2
ENST00000602964.1 ENST00000519898.1 |
MIR143HG
|
MIR143 host gene (non-protein coding) |
| chr17_+_11501748 | 0.07 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr1_+_151009054 | 0.07 |
ENST00000295294.7
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr1_+_151009035 | 0.07 |
ENST00000368931.3
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr3_+_89156674 | 0.06 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr14_-_74960030 | 0.06 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr19_-_6767516 | 0.06 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr11_+_74303575 | 0.06 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr1_+_100503643 | 0.06 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr4_-_85419603 | 0.06 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr7_+_48211048 | 0.06 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr10_-_98273668 | 0.05 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr7_+_23637763 | 0.05 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr11_+_8704748 | 0.05 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr10_+_90660832 | 0.05 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr7_+_102389434 | 0.05 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr11_+_72525353 | 0.04 |
ENST00000321297.5
ENST00000534905.1 ENST00000540567.1 |
ATG16L2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr11_-_59951889 | 0.04 |
ENST00000532169.1
ENST00000534596.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr1_+_35220613 | 0.04 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr19_-_6767431 | 0.04 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr10_+_52152766 | 0.04 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr1_+_171107241 | 0.03 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr3_+_89156799 | 0.03 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr4_-_99851766 | 0.03 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr12_-_56321397 | 0.03 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr9_-_4666421 | 0.03 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr11_-_119252359 | 0.03 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr11_+_63273811 | 0.03 |
ENST00000340246.5
|
LGALS12
|
lectin, galactoside-binding, soluble, 12 |
| chr7_+_129984630 | 0.03 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr18_-_47018769 | 0.03 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr6_+_140175987 | 0.03 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr2_+_197577841 | 0.03 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr4_-_36246060 | 0.03 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_+_60145967 | 0.02 |
ENST00000534016.1
|
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr16_+_31724552 | 0.02 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr9_-_28026318 | 0.02 |
ENST00000308675.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr21_-_32253874 | 0.02 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr12_+_72080253 | 0.02 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr15_-_66797172 | 0.02 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chrX_-_122866874 | 0.01 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr12_-_123215306 | 0.01 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr7_-_142247606 | 0.01 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr10_+_63422695 | 0.01 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr22_-_36236623 | 0.01 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_35089722 | 0.01 |
ENST00000511486.1
ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR
|
prolactin receptor |
| chr15_+_93015151 | 0.01 |
ENST00000556865.1
|
C15orf32
|
chromosome 15 open reading frame 32 |
| chr12_-_7077125 | 0.01 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr1_+_46016703 | 0.01 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr12_+_14572070 | 0.00 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr16_-_30134524 | 0.00 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 2.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.5 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 2.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.6 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:0048633 | cell-cell signaling involved in cell fate commitment(GO:0045168) negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 2.3 | GO:0030224 | monocyte differentiation(GO:0030224) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.9 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.8 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0090545 | Sin3 complex(GO:0016580) NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0030877 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |