Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for ZBTB18
Z-value: 0.66
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
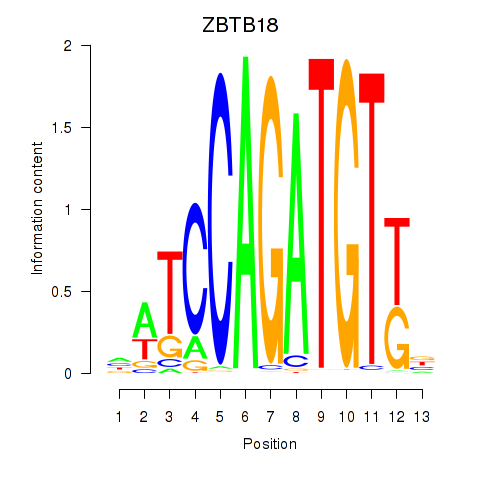
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | -0.56 | 3.8e-03 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_152214098 | 3.31 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr5_+_118690466 | 2.52 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_+_77020325 | 1.75 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 1.75 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr15_+_67418047 | 1.47 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_+_201994042 | 1.24 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_108994466 | 1.11 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr17_+_77020146 | 1.05 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_+_112563335 | 0.90 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_112563303 | 0.89 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_+_150151824 | 0.80 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chrX_-_154563889 | 0.74 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr2_-_145278475 | 0.72 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_49962495 | 0.69 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr3_-_37216055 | 0.63 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr13_+_97874574 | 0.61 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_-_57089671 | 0.61 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr17_+_32582293 | 0.59 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr2_-_152146385 | 0.57 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr9_+_100174344 | 0.55 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr9_+_118916082 | 0.55 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr9_+_100174232 | 0.53 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr12_+_26126681 | 0.53 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_-_89399845 | 0.52 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chrX_+_64808248 | 0.52 |
ENST00000609672.1
|
MSN
|
moesin |
| chr7_+_100770328 | 0.51 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr2_-_230096756 | 0.50 |
ENST00000354069.6
|
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr6_+_155537771 | 0.49 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr16_+_24266874 | 0.48 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr19_+_11649532 | 0.48 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chr11_+_71249071 | 0.47 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr17_+_32612687 | 0.40 |
ENST00000305869.3
|
CCL11
|
chemokine (C-C motif) ligand 11 |
| chr14_-_61748550 | 0.39 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr4_-_105416039 | 0.39 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr7_-_122339162 | 0.37 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr2_+_191045562 | 0.36 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr3_-_44552094 | 0.36 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr5_-_151304337 | 0.36 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr9_+_72658490 | 0.35 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr15_-_55611306 | 0.34 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_+_97733786 | 0.33 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chrX_-_111923145 | 0.32 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr22_-_37823468 | 0.32 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr17_+_32597232 | 0.32 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr2_-_89597542 | 0.31 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr5_+_140792614 | 0.30 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chrX_+_48242863 | 0.30 |
ENST00000376886.2
ENST00000375517.3 |
SSX4
|
synovial sarcoma, X breakpoint 4 |
| chr11_-_5537920 | 0.29 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr15_+_51669444 | 0.29 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chrX_-_19688475 | 0.28 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr20_+_57875658 | 0.28 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chrX_-_110513703 | 0.27 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr11_+_94227129 | 0.27 |
ENST00000540349.1
ENST00000535502.1 ENST00000545130.1 ENST00000544253.1 ENST00000541144.1 |
ANKRD49
|
ankyrin repeat domain 49 |
| chr2_+_89923550 | 0.27 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr7_-_71868354 | 0.27 |
ENST00000412588.1
|
CALN1
|
calneuron 1 |
| chr2_-_7005785 | 0.26 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr11_+_5474638 | 0.26 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr1_-_2458026 | 0.25 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr6_-_163148700 | 0.25 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr2_-_74692473 | 0.25 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr16_-_30394143 | 0.25 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr19_+_39903185 | 0.24 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr9_+_116263778 | 0.23 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_+_9005917 | 0.23 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chrX_+_150151752 | 0.23 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr4_+_15376165 | 0.23 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr14_+_67707826 | 0.23 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr9_+_116263639 | 0.23 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chrY_-_21239221 | 0.23 |
ENST00000447937.1
ENST00000331787.2 |
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr12_+_1738363 | 0.23 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr11_-_34379546 | 0.22 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr10_+_105253661 | 0.22 |
ENST00000369780.4
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr16_+_28914680 | 0.22 |
ENST00000564112.1
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_+_273038 | 0.22 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr8_+_104831472 | 0.22 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_26737253 | 0.21 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr3_-_160167508 | 0.21 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr16_-_31076332 | 0.20 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr11_+_128562372 | 0.20 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_42619070 | 0.20 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chrX_+_103031758 | 0.20 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr5_-_149324306 | 0.19 |
ENST00000255266.5
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr5_+_159895275 | 0.19 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr9_+_87283430 | 0.19 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_+_59824127 | 0.19 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr17_-_15168624 | 0.19 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr1_-_153518270 | 0.18 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr2_-_89417335 | 0.18 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr11_+_65405556 | 0.18 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr12_-_54785074 | 0.18 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chrX_+_103031421 | 0.17 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr7_-_37024665 | 0.17 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr6_+_163148973 | 0.17 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr15_+_31619013 | 0.16 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr12_-_54785054 | 0.16 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_26737292 | 0.16 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr1_-_217262969 | 0.16 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr21_-_34185944 | 0.16 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr19_+_42806250 | 0.15 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr5_+_132009675 | 0.15 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr11_+_32112431 | 0.14 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr11_+_59824060 | 0.14 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr11_+_15136462 | 0.14 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr16_+_6069586 | 0.14 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_+_140782351 | 0.14 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chrX_-_30877837 | 0.13 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr11_-_40315640 | 0.13 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr4_+_156824840 | 0.13 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr7_+_62809239 | 0.13 |
ENST00000456890.1
|
AC006455.1
|
AC006455.1 |
| chr4_+_75311019 | 0.13 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr2_-_163099885 | 0.12 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr19_+_9251052 | 0.12 |
ENST00000247956.6
ENST00000360385.3 |
ZNF317
|
zinc finger protein 317 |
| chrX_-_48056199 | 0.12 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chrX_-_77225135 | 0.12 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr12_+_2162447 | 0.12 |
ENST00000335762.5
ENST00000399655.1 |
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr11_-_62323702 | 0.12 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr2_-_163100045 | 0.11 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr2_+_90139056 | 0.11 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr17_-_19651654 | 0.11 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chrX_+_153770421 | 0.10 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr20_+_57875758 | 0.10 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr2_+_219247021 | 0.10 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr17_-_19651668 | 0.10 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_-_217262933 | 0.10 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr17_+_19912640 | 0.10 |
ENST00000395527.4
ENST00000583482.2 ENST00000583528.1 ENST00000583463.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_-_43559055 | 0.10 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr4_-_111563076 | 0.10 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr8_-_49834299 | 0.09 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr20_+_30467600 | 0.09 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr6_-_39197226 | 0.09 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr21_-_35883541 | 0.08 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr2_+_113885138 | 0.08 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_-_163148780 | 0.08 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr17_+_48638371 | 0.08 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr8_-_49833978 | 0.08 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr17_-_62050278 | 0.07 |
ENST00000578147.1
ENST00000435607.1 |
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit |
| chr19_+_50936142 | 0.07 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr11_-_128894053 | 0.06 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr19_-_52148798 | 0.06 |
ENST00000534261.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr9_+_97488939 | 0.06 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr6_+_160542821 | 0.06 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_+_32608566 | 0.06 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr3_+_121311966 | 0.06 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr1_-_156647189 | 0.06 |
ENST00000368223.3
|
NES
|
nestin |
| chr12_-_56122124 | 0.06 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr2_-_225434538 | 0.06 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr11_+_76494253 | 0.05 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr3_-_52273098 | 0.05 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr15_+_49715293 | 0.05 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr4_+_47487285 | 0.05 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr10_-_71332994 | 0.05 |
ENST00000242462.4
|
NEUROG3
|
neurogenin 3 |
| chr15_-_31453157 | 0.05 |
ENST00000559177.1
ENST00000558445.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr13_+_113777105 | 0.04 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr4_+_88571429 | 0.04 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr7_+_95115210 | 0.04 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr10_+_98064085 | 0.04 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr1_-_99470558 | 0.04 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr19_+_48673949 | 0.04 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr7_-_154863264 | 0.04 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr17_-_41174424 | 0.04 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr12_-_56122426 | 0.04 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr11_+_63870660 | 0.03 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr7_-_156803329 | 0.03 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr12_-_56121580 | 0.03 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr12_-_48119340 | 0.03 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr12_-_56122220 | 0.03 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr3_-_160167301 | 0.03 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr1_+_38022572 | 0.02 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr1_-_11907829 | 0.02 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr20_+_56725952 | 0.02 |
ENST00000371168.3
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr6_-_74363803 | 0.02 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr1_-_99470368 | 0.02 |
ENST00000263177.4
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr10_-_123357598 | 0.02 |
ENST00000358487.5
ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr12_-_48119301 | 0.01 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr6_+_160542870 | 0.01 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_+_53527854 | 0.01 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr17_+_42385927 | 0.01 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr19_-_15918936 | 0.01 |
ENST00000334920.2
|
OR10H1
|
olfactory receptor, family 10, subfamily H, member 1 |
| chr15_+_92937144 | 0.00 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.6 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 0.5 | GO:0001300 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 1.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.3 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.7 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 1.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 1.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 1.8 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.5 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 2.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 4.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0043219 | myelin sheath adaxonal region(GO:0035749) lateral loop(GO:0043219) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 3.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 2.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 1.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 4.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 1.2 | GO:0097200 | death receptor binding(GO:0005123) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 1.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 4.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |