Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
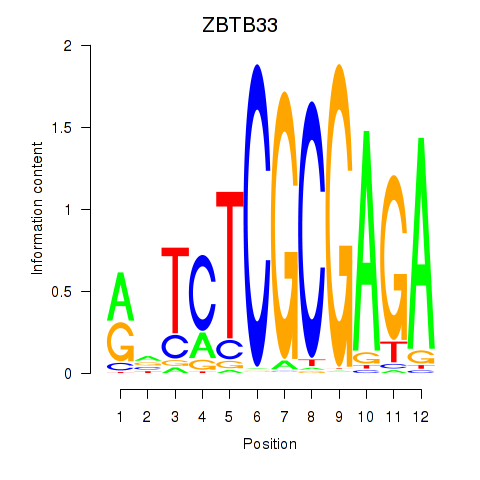
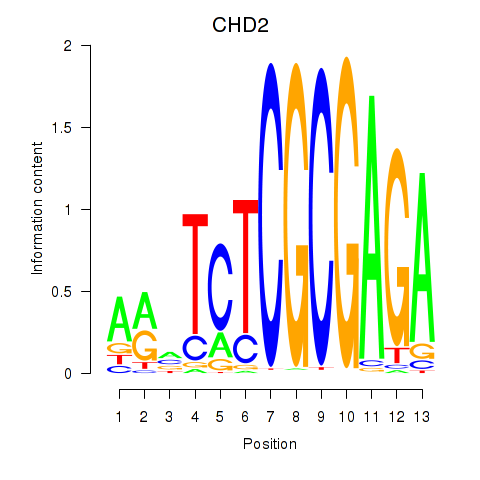
Results for ZBTB33_CHD2
Z-value: 1.39
Transcription factors associated with ZBTB33_CHD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB33
|
ENSG00000177485.6 | zinc finger and BTB domain containing 33 |
|
CHD2
|
ENSG00000173575.14 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CHD2 | hg19_v2_chr15_+_93443419_93443580 | -0.24 | 2.6e-01 | Click! |
| ZBTB33 | hg19_v2_chrX_+_119384607_119384720 | 0.11 | 6.0e-01 | Click! |
Activity profile of ZBTB33_CHD2 motif
Sorted Z-values of ZBTB33_CHD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_7761301 | 5.59 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr12_+_27396901 | 3.83 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr3_+_8543393 | 3.30 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_+_8543561 | 3.29 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr13_+_115047097 | 3.29 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr13_+_115047053 | 3.10 |
ENST00000375299.3
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr13_+_42846272 | 2.92 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr5_+_176449726 | 2.74 |
ENST00000261948.4
ENST00000511834.1 ENST00000503039.1 |
ZNF346
|
zinc finger protein 346 |
| chr5_+_176449684 | 2.46 |
ENST00000506693.1
ENST00000358149.3 ENST00000512315.1 ENST00000503425.1 |
ZNF346
|
zinc finger protein 346 |
| chr6_-_146285455 | 2.43 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr3_+_8543533 | 2.29 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr6_-_146285221 | 2.29 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr1_+_104068562 | 2.20 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr12_-_27091183 | 2.14 |
ENST00000544548.1
ENST00000261191.7 ENST00000537336.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr1_-_92371839 | 2.05 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr12_-_27090896 | 2.02 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr8_+_37963311 | 1.90 |
ENST00000428278.2
ENST00000521652.1 |
ASH2L
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr3_-_18466026 | 1.89 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr8_+_37963011 | 1.88 |
ENST00000250635.7
ENST00000517719.1 ENST00000545394.1 |
ASH2L
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr1_-_78148324 | 1.85 |
ENST00000370801.3
ENST00000433749.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr13_-_21750659 | 1.84 |
ENST00000400018.3
ENST00000314759.5 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr1_+_104068312 | 1.82 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr10_-_127408011 | 1.73 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr9_-_125667618 | 1.71 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr17_-_7761172 | 1.68 |
ENST00000333775.5
ENST00000575771.1 |
LSMD1
|
LSM domain containing 1 |
| chr12_-_80328700 | 1.67 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr17_-_53046058 | 1.64 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr19_-_37263693 | 1.64 |
ENST00000591344.1
|
ZNF850
|
zinc finger protein 850 |
| chr2_-_677369 | 1.64 |
ENST00000281017.3
|
TMEM18
|
transmembrane protein 18 |
| chr3_-_160117301 | 1.60 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr15_-_56035177 | 1.59 |
ENST00000389286.4
ENST00000561292.1 |
PRTG
|
protogenin |
| chr12_+_27091426 | 1.58 |
ENST00000546072.1
ENST00000327214.5 |
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr12_+_27091316 | 1.52 |
ENST00000229395.3
|
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr7_-_129845313 | 1.52 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr2_+_170683979 | 1.51 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr18_+_59854480 | 1.43 |
ENST00000256858.6
ENST00000398130.2 |
KIAA1468
|
KIAA1468 |
| chr20_+_17949544 | 1.39 |
ENST00000377710.5
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr5_-_89705537 | 1.38 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr17_+_4843679 | 1.37 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr11_-_118436606 | 1.37 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr2_+_170683942 | 1.37 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr11_-_118436707 | 1.34 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr20_+_17949684 | 1.34 |
ENST00000377709.1
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr6_-_116575226 | 1.33 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr3_+_42642106 | 1.32 |
ENST00000232978.8
|
NKTR
|
natural killer-tumor recognition sequence |
| chr16_+_14165160 | 1.29 |
ENST00000574998.1
ENST00000571589.1 ENST00000574045.1 |
MKL2
|
MKL/myocardin-like 2 |
| chr6_+_35995531 | 1.28 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr20_+_17949724 | 1.27 |
ENST00000377704.4
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr6_+_35995488 | 1.27 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr2_-_55844720 | 1.24 |
ENST00000345102.5
ENST00000272313.5 ENST00000407823.3 |
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
| chr18_-_59854203 | 1.23 |
ENST00000589339.1
ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr9_-_125667494 | 1.21 |
ENST00000335387.5
ENST00000357244.2 ENST00000373665.2 |
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr3_-_165555200 | 1.19 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_-_10249990 | 1.19 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chr11_-_94227029 | 1.18 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr4_+_17812525 | 1.17 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr3_+_160117418 | 1.17 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr22_-_21356375 | 1.17 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr15_+_78730531 | 1.16 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr4_-_17812309 | 1.16 |
ENST00000382247.1
ENST00000536863.1 |
DCAF16
|
DDB1 and CUL4 associated factor 16 |
| chr6_+_71122974 | 1.16 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr10_-_64576105 | 1.14 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr1_+_231473743 | 1.14 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr12_-_80328949 | 1.14 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr3_-_93781750 | 1.11 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr1_+_26146397 | 1.09 |
ENST00000374303.2
ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr3_+_32280159 | 1.09 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr12_+_123237321 | 1.08 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr13_-_22178284 | 1.06 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr1_+_174969262 | 1.06 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr5_-_73937244 | 1.04 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr1_+_26146674 | 1.03 |
ENST00000525713.1
ENST00000374301.3 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr9_+_26956371 | 1.03 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr3_+_160117087 | 1.03 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr5_-_72861484 | 1.01 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr12_+_123319973 | 1.01 |
ENST00000253083.4
|
HIP1R
|
huntingtin interacting protein 1 related |
| chr17_+_4843654 | 0.99 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167 |
| chr17_+_4843303 | 0.99 |
ENST00000571816.1
|
RNF167
|
ring finger protein 167 |
| chr11_+_93474786 | 0.98 |
ENST00000331239.4
ENST00000533585.1 ENST00000528099.1 ENST00000354421.3 ENST00000540113.1 ENST00000530620.1 ENST00000527003.1 ENST00000531650.1 ENST00000530279.1 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr19_+_42364313 | 0.98 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr4_+_25378912 | 0.96 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr17_-_61920280 | 0.95 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr17_-_7761256 | 0.94 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr13_-_41768654 | 0.93 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr12_-_29534074 | 0.92 |
ENST00000546839.1
ENST00000360150.4 ENST00000552155.1 ENST00000550353.1 ENST00000548441.1 ENST00000552132.1 |
ERGIC2
|
ERGIC and golgi 2 |
| chr7_+_2443202 | 0.91 |
ENST00000258711.6
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr2_-_43823093 | 0.91 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr6_+_25279651 | 0.91 |
ENST00000329474.6
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr17_+_34900737 | 0.90 |
ENST00000304718.4
ENST00000485685.2 |
GGNBP2
|
gametogenetin binding protein 2 |
| chr15_+_44580899 | 0.89 |
ENST00000559222.1
ENST00000299957.6 |
CASC4
|
cancer susceptibility candidate 4 |
| chr21_-_34144157 | 0.89 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr4_-_178363581 | 0.88 |
ENST00000264595.2
|
AGA
|
aspartylglucosaminidase |
| chr6_+_71123107 | 0.87 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr17_+_4843413 | 0.87 |
ENST00000572430.1
ENST00000262482.6 |
RNF167
|
ring finger protein 167 |
| chr6_+_2765595 | 0.87 |
ENST00000380773.4
ENST00000380771.4 |
WRNIP1
|
Werner helicase interacting protein 1 |
| chr4_+_25378826 | 0.87 |
ENST00000315368.3
|
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr14_+_24867992 | 0.86 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr10_-_126847276 | 0.86 |
ENST00000531469.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_28052456 | 0.86 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr9_+_139971921 | 0.85 |
ENST00000409858.3
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
| chr19_+_42364460 | 0.85 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr10_+_54074033 | 0.83 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_+_35995552 | 0.81 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr10_-_77161650 | 0.80 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr18_+_77623668 | 0.80 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr15_+_44580955 | 0.80 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr2_-_201753980 | 0.78 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr1_+_100316041 | 0.78 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr7_-_75677251 | 0.78 |
ENST00000431581.1
ENST00000359697.3 ENST00000451157.1 ENST00000340062.5 ENST00000360591.3 ENST00000248600.1 |
STYXL1
|
serine/threonine/tyrosine interacting-like 1 |
| chr10_+_127408263 | 0.77 |
ENST00000337623.3
|
C10orf137
|
erythroid differentiation regulatory factor 1 |
| chr2_-_222436988 | 0.76 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr1_+_100315613 | 0.76 |
ENST00000361915.3
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr1_-_174992544 | 0.76 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr7_-_35734730 | 0.76 |
ENST00000396081.1
ENST00000311350.3 |
HERPUD2
|
HERPUD family member 2 |
| chr4_+_71768043 | 0.75 |
ENST00000502869.1
ENST00000309395.2 ENST00000396051.2 |
MOB1B
|
MOB kinase activator 1B |
| chr19_+_42363917 | 0.74 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr4_+_144434584 | 0.74 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr13_-_79979952 | 0.73 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr2_+_203499901 | 0.72 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr2_+_214149113 | 0.71 |
ENST00000331683.5
ENST00000432529.2 ENST00000413312.1 ENST00000272898.7 ENST00000447990.1 |
SPAG16
|
sperm associated antigen 16 |
| chr15_-_73925651 | 0.71 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr3_-_101232019 | 0.71 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr3_+_108308513 | 0.70 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr4_+_120133791 | 0.70 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr11_-_71791726 | 0.70 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_-_133263893 | 0.70 |
ENST00000535270.1
ENST00000320574.5 |
POLE
|
polymerase (DNA directed), epsilon, catalytic subunit |
| chr2_+_120517174 | 0.70 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr6_+_3849584 | 0.70 |
ENST00000380274.1
ENST00000380272.3 |
FAM50B
|
family with sequence similarity 50, member B |
| chr2_-_39664405 | 0.69 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr13_+_100741269 | 0.69 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chrX_-_13752675 | 0.69 |
ENST00000380579.1
ENST00000458511.2 ENST00000519885.1 ENST00000358231.5 ENST00000518847.1 ENST00000453655.2 ENST00000359680.5 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr15_-_34880646 | 0.68 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr11_+_47291645 | 0.68 |
ENST00000395336.3
ENST00000402192.2 |
MADD
|
MAP-kinase activating death domain |
| chr8_+_143808605 | 0.67 |
ENST00000336138.3
|
THEM6
|
thioesterase superfamily member 6 |
| chr1_-_45672221 | 0.67 |
ENST00000359600.5
|
ZSWIM5
|
zinc finger, SWIM-type containing 5 |
| chr9_-_123605177 | 0.66 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr19_+_46002868 | 0.66 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr5_+_36152163 | 0.65 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr13_-_79979919 | 0.65 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr16_-_80926457 | 0.65 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr20_+_49126881 | 0.65 |
ENST00000371621.3
ENST00000541713.1 |
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr15_-_48470544 | 0.65 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr14_+_61788429 | 0.65 |
ENST00000332981.5
|
PRKCH
|
protein kinase C, eta |
| chr15_-_48470558 | 0.65 |
ENST00000324324.7
|
MYEF2
|
myelin expression factor 2 |
| chr11_-_71791435 | 0.64 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr4_+_128886532 | 0.64 |
ENST00000444616.1
ENST00000388795.5 |
C4orf29
|
chromosome 4 open reading frame 29 |
| chr20_-_17949100 | 0.64 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr12_+_102513950 | 0.64 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr11_-_71791518 | 0.64 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr5_-_73936451 | 0.64 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr2_-_20101701 | 0.63 |
ENST00000402414.1
ENST00000333610.3 |
TTC32
|
tetratricopeptide repeat domain 32 |
| chr12_+_56661033 | 0.63 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr2_-_157189180 | 0.63 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_16761678 | 0.62 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr4_+_128886424 | 0.62 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr1_-_70820357 | 0.62 |
ENST00000370944.4
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr11_+_93474757 | 0.62 |
ENST00000528288.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr4_-_68411275 | 0.62 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr5_+_36152091 | 0.61 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr10_-_7829909 | 0.61 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr8_-_145980968 | 0.60 |
ENST00000292562.7
|
ZNF251
|
zinc finger protein 251 |
| chr1_-_6295975 | 0.60 |
ENST00000343813.5
ENST00000362035.3 |
ICMT
|
isoprenylcysteine carboxyl methyltransferase |
| chr7_-_150038704 | 0.60 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr5_+_138629337 | 0.60 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr8_+_95835438 | 0.60 |
ENST00000521860.1
ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8
|
integrator complex subunit 8 |
| chr1_+_24742264 | 0.60 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr3_-_137893721 | 0.59 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr4_-_110624564 | 0.58 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr4_-_90758227 | 0.58 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_-_7760779 | 0.58 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr2_+_189156389 | 0.57 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_+_55162453 | 0.57 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chrX_+_48554986 | 0.57 |
ENST00000376687.3
ENST00000453214.2 |
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr19_-_53193731 | 0.57 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr2_-_43823119 | 0.57 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr1_+_111991474 | 0.56 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr11_+_11863579 | 0.56 |
ENST00000399455.2
|
USP47
|
ubiquitin specific peptidase 47 |
| chr2_+_189156721 | 0.56 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_9563697 | 0.56 |
ENST00000238112.3
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr3_+_169684553 | 0.56 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr12_-_124018252 | 0.56 |
ENST00000376874.4
|
RILPL1
|
Rab interacting lysosomal protein-like 1 |
| chr2_+_189156586 | 0.56 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_+_74723031 | 0.55 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr14_+_61447832 | 0.55 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr13_-_37633567 | 0.55 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr5_+_118407053 | 0.55 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr9_+_86595626 | 0.54 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr3_-_183735731 | 0.54 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_-_39074479 | 0.54 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr2_+_9564013 | 0.54 |
ENST00000460593.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr4_+_170541660 | 0.52 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr21_+_35445827 | 0.51 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr7_-_158497431 | 0.51 |
ENST00000449727.2
ENST00000409339.3 ENST00000409423.1 ENST00000356309.3 |
NCAPG2
|
non-SMC condensin II complex, subunit G2 |
| chr16_-_18801643 | 0.51 |
ENST00000322989.4
ENST00000563390.1 |
RPS15A
|
ribosomal protein S15a |
| chr5_+_134181755 | 0.51 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr9_+_35792151 | 0.51 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr6_+_130686856 | 0.51 |
ENST00000296978.3
|
TMEM200A
|
transmembrane protein 200A |
| chr5_+_139493665 | 0.50 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr15_+_22833395 | 0.50 |
ENST00000283645.4
|
TUBGCP5
|
tubulin, gamma complex associated protein 5 |
| chr21_+_34638656 | 0.49 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB33_CHD2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.7 | 3.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 2.0 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.6 | 2.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.6 | 2.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 2.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 1.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.1 | GO:0035283 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 1.5 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.4 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 8.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 2.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 2.9 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 0.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 1.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 0.8 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 0.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 2.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 1.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 0.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.7 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 1.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.6 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 0.9 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.2 | 0.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 1.0 | GO:0015853 | adenine transport(GO:0015853) |
| 0.2 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.0 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.4 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.5 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.5 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 1.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.4 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 1.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.3 | GO:1904437 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 5.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.7 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 | 0.4 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 3.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.4 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.1 | 0.5 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 1.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.7 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.6 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.6 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 1.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.6 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 3.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.3 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.3 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 1.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 2.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.0 | 0.2 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0003163 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:1902739 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.1 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 4.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.9 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 3.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.7 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 3.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 1.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.5 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 1.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.0 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.3 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.8 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.8 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.6 | 2.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 3.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 4.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 2.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 0.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 2.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 5.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.5 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 4.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 3.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0035631 | IkappaB kinase complex(GO:0008385) CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 3.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.4 | 1.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 1.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 4.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.3 | 1.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.3 | 3.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 2.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 3.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 0.5 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.2 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 6.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.1 | 0.6 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 2.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.7 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 4.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0016662 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0034711 | activin-activated receptor activity(GO:0017002) inhibin binding(GO:0034711) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.9 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 8.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 9.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 5.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.1 | REACTOME DEVELOPMENTAL BIOLOGY | Genes involved in Developmental Biology |
| 0.1 | 2.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 4.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |