Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
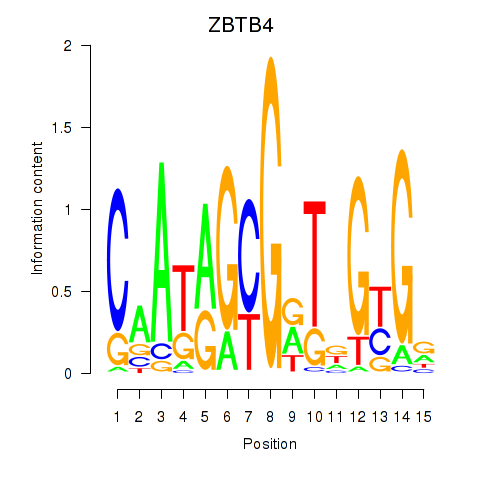
Results for ZBTB4
Z-value: 0.37
Transcription factors associated with ZBTB4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB4
|
ENSG00000174282.7 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB4 | hg19_v2_chr17_-_7387524_7387597, hg19_v2_chr17_-_7382834_7382855 | -0.32 | 1.2e-01 | Click! |
Activity profile of ZBTB4 motif
Sorted Z-values of ZBTB4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_72385437 | 1.19 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr1_+_169075554 | 0.84 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr10_-_101380121 | 0.81 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr17_-_40264692 | 0.50 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr11_+_86511549 | 0.44 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chrX_-_68385354 | 0.43 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chrX_-_68385274 | 0.43 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr13_-_44361025 | 0.39 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr13_-_33780133 | 0.38 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr15_-_40212363 | 0.37 |
ENST00000299092.3
|
GPR176
|
G protein-coupled receptor 176 |
| chr15_-_40213080 | 0.35 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr16_-_87903079 | 0.32 |
ENST00000261622.4
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr1_-_203055129 | 0.31 |
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4) |
| chr11_+_62623544 | 0.28 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr3_-_192635943 | 0.28 |
ENST00000392452.2
|
MB21D2
|
Mab-21 domain containing 2 |
| chr11_+_62623512 | 0.27 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr10_+_37414785 | 0.26 |
ENST00000374660.1
ENST00000602533.1 ENST00000361713.1 |
ANKRD30A
|
ankyrin repeat domain 30A |
| chr5_-_146889619 | 0.26 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr14_-_85996332 | 0.26 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr16_-_79634595 | 0.25 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr16_-_89556942 | 0.23 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr8_+_94929077 | 0.23 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_+_86511569 | 0.22 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr17_-_30185946 | 0.22 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr22_+_29279552 | 0.21 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr4_-_7069760 | 0.21 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr1_+_166808692 | 0.21 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr7_-_18067478 | 0.21 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr14_-_69445793 | 0.21 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr14_+_74417192 | 0.21 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr8_+_94929110 | 0.20 |
ENST00000520728.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_-_71581377 | 0.20 |
ENST00000276590.4
ENST00000522447.1 |
LACTB2
|
lactamase, beta 2 |
| chr5_-_96143796 | 0.20 |
ENST00000296754.3
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr11_+_33902189 | 0.19 |
ENST00000330381.2
|
AC132216.1
|
HCG1785179; PRO1787; Uncharacterized protein |
| chr6_-_136847610 | 0.19 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr6_+_159290917 | 0.19 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr11_-_63439381 | 0.19 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr11_+_65082289 | 0.19 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr1_-_156786530 | 0.18 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr9_+_4792971 | 0.18 |
ENST00000381732.3
ENST00000442869.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr22_-_50970919 | 0.17 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chrX_-_71526813 | 0.17 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr5_-_115177247 | 0.17 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr6_+_31582961 | 0.17 |
ENST00000376059.3
ENST00000337917.7 |
AIF1
|
allograft inflammatory factor 1 |
| chr3_-_167452703 | 0.17 |
ENST00000497056.2
ENST00000473645.2 |
PDCD10
|
programmed cell death 10 |
| chr1_-_151254362 | 0.17 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr16_-_11370330 | 0.17 |
ENST00000241808.4
ENST00000435245.2 |
PRM2
|
protamine 2 |
| chr2_+_192110199 | 0.17 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr7_+_87257854 | 0.16 |
ENST00000394654.3
|
RUNDC3B
|
RUN domain containing 3B |
| chr11_+_120973375 | 0.16 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr13_+_24734880 | 0.16 |
ENST00000382095.4
|
SPATA13
|
spermatogenesis associated 13 |
| chr16_-_30042580 | 0.16 |
ENST00000380495.4
|
FAM57B
|
family with sequence similarity 57, member B |
| chr19_+_17830051 | 0.16 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr15_-_98417780 | 0.15 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr11_+_125462690 | 0.15 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr3_-_167452614 | 0.15 |
ENST00000392750.2
ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10
|
programmed cell death 10 |
| chr17_-_5389477 | 0.15 |
ENST00000572834.1
ENST00000570848.1 ENST00000571971.1 ENST00000158771.4 |
DERL2
|
derlin 2 |
| chr2_+_192109911 | 0.15 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr17_-_30185971 | 0.15 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr19_+_6135646 | 0.15 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr17_-_48943706 | 0.15 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr6_-_114664180 | 0.15 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr8_+_94929168 | 0.14 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr22_-_50970506 | 0.14 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr4_+_71554196 | 0.14 |
ENST00000254803.2
|
UTP3
|
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr3_+_113666748 | 0.14 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr10_+_123922941 | 0.14 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_33283754 | 0.14 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chrX_-_74145273 | 0.14 |
ENST00000055682.6
|
KIAA2022
|
KIAA2022 |
| chr16_+_640201 | 0.13 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr3_+_40518599 | 0.13 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr19_-_10530784 | 0.13 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr7_-_5463175 | 0.13 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr17_-_2996290 | 0.13 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr15_-_75660919 | 0.13 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr17_-_30186328 | 0.13 |
ENST00000302362.6
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr13_-_24007815 | 0.13 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr17_-_56595196 | 0.12 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr1_-_151299842 | 0.12 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr1_+_200842083 | 0.12 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr3_-_119813264 | 0.12 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr16_-_30538079 | 0.12 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr17_+_54671047 | 0.12 |
ENST00000332822.4
|
NOG
|
noggin |
| chr9_-_131709858 | 0.11 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr1_+_110163709 | 0.11 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr9_-_80263220 | 0.11 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr17_+_34058639 | 0.11 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr14_-_74417096 | 0.11 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr14_-_69445968 | 0.11 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr20_+_61147651 | 0.11 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr18_-_55253871 | 0.11 |
ENST00000382873.3
|
FECH
|
ferrochelatase |
| chr19_+_39421556 | 0.11 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr8_-_143851203 | 0.11 |
ENST00000521396.1
ENST00000317543.7 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr19_+_19144384 | 0.10 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr12_+_5541267 | 0.10 |
ENST00000423158.3
|
NTF3
|
neurotrophin 3 |
| chr7_-_128694927 | 0.10 |
ENST00000471166.1
ENST00000265388.5 |
TNPO3
|
transportin 3 |
| chr6_+_31583761 | 0.10 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr11_-_73471655 | 0.10 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr7_-_44613494 | 0.10 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr10_-_103599591 | 0.10 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr17_+_5389605 | 0.10 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr19_+_47759716 | 0.09 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr3_-_57583130 | 0.09 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr19_-_50316517 | 0.09 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr11_-_61735029 | 0.09 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chrX_-_47509887 | 0.09 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr9_-_128246769 | 0.09 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_-_100487280 | 0.08 |
ENST00000388761.2
|
UFSP1
|
UFM1-specific peptidase 1 (non-functional) |
| chr19_+_10828795 | 0.08 |
ENST00000389253.4
ENST00000355667.6 ENST00000408974.4 |
DNM2
|
dynamin 2 |
| chr4_-_156298087 | 0.08 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr6_-_48036363 | 0.08 |
ENST00000543600.1
ENST00000398738.2 ENST00000339488.4 |
PTCHD4
|
patched domain containing 4 |
| chr1_-_160549235 | 0.08 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr4_-_156297949 | 0.08 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr19_+_36024310 | 0.08 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr14_-_35099377 | 0.08 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr11_+_107799118 | 0.08 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr3_-_57583052 | 0.08 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr19_+_10531150 | 0.08 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr19_+_10828724 | 0.08 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr14_-_35099315 | 0.08 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr17_+_72270380 | 0.08 |
ENST00000582036.1
ENST00000307504.5 |
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr2_+_152266392 | 0.08 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr22_-_50970566 | 0.07 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr16_-_22385901 | 0.07 |
ENST00000268383.2
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa |
| chr8_+_57124245 | 0.07 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chrX_+_147582130 | 0.07 |
ENST00000370460.2
ENST00000370457.5 |
AFF2
|
AF4/FMR2 family, member 2 |
| chr11_-_63439174 | 0.07 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr7_+_87257701 | 0.07 |
ENST00000338056.3
ENST00000493037.1 |
RUNDC3B
|
RUN domain containing 3B |
| chr3_-_57583185 | 0.07 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr14_+_55494323 | 0.06 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr2_+_155554797 | 0.06 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr1_+_10534944 | 0.06 |
ENST00000356607.4
ENST00000538836.1 ENST00000491661.2 |
PEX14
|
peroxisomal biogenesis factor 14 |
| chr9_-_136242956 | 0.06 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr16_+_640055 | 0.06 |
ENST00000568586.1
ENST00000538492.1 ENST00000248139.3 |
RAB40C
|
RAB40C, member RAS oncogene family |
| chr4_-_113437328 | 0.06 |
ENST00000313341.3
|
NEUROG2
|
neurogenin 2 |
| chr1_-_32403370 | 0.06 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_165738072 | 0.06 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr4_+_4388805 | 0.06 |
ENST00000504171.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr15_-_44116873 | 0.06 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr1_-_38325256 | 0.06 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr14_+_55493920 | 0.06 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr14_+_74416989 | 0.06 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr17_+_45727204 | 0.06 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr2_-_180871780 | 0.06 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr19_+_1275917 | 0.06 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr2_+_232651124 | 0.06 |
ENST00000350033.3
ENST00000412591.1 ENST00000410017.1 ENST00000373608.3 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr14_+_23305760 | 0.06 |
ENST00000311852.6
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr12_+_6561190 | 0.05 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr2_-_207024233 | 0.05 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr20_-_43438912 | 0.05 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr6_+_158244223 | 0.05 |
ENST00000392185.3
|
SNX9
|
sorting nexin 9 |
| chr19_+_52074502 | 0.05 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr8_-_74205851 | 0.05 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr8_-_29208183 | 0.05 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr9_-_130497565 | 0.05 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr9_+_33290491 | 0.05 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr7_+_102937869 | 0.05 |
ENST00000249269.4
ENST00000428154.1 ENST00000420236.2 |
PMPCB
|
peptidase (mitochondrial processing) beta |
| chr19_-_20748541 | 0.05 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr17_-_19265855 | 0.04 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr19_-_50316489 | 0.04 |
ENST00000533418.1
|
FUZ
|
fuzzy planar cell polarity protein |
| chr11_-_63439013 | 0.04 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr17_+_55334364 | 0.04 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr12_-_91348949 | 0.04 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr19_+_20959142 | 0.04 |
ENST00000344519.8
|
ZNF66
|
zinc finger protein 66 |
| chr9_-_71629010 | 0.04 |
ENST00000377276.2
|
PRKACG
|
protein kinase, cAMP-dependent, catalytic, gamma |
| chr2_-_201936302 | 0.04 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr8_-_16859690 | 0.04 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr19_-_50316423 | 0.04 |
ENST00000528094.1
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr1_+_151043070 | 0.04 |
ENST00000368918.3
ENST00000368917.1 |
GABPB2
|
GA binding protein transcription factor, beta subunit 2 |
| chr4_-_83295103 | 0.04 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr12_-_49525175 | 0.04 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr2_-_191399426 | 0.04 |
ENST00000409150.3
|
TMEM194B
|
transmembrane protein 194B |
| chr10_-_53459319 | 0.03 |
ENST00000331173.4
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr2_-_203735976 | 0.03 |
ENST00000435143.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr14_-_50559361 | 0.03 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr22_+_21987005 | 0.03 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr12_+_52282001 | 0.03 |
ENST00000340970.4
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr3_+_188889737 | 0.03 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr19_+_15121532 | 0.03 |
ENST00000292574.3
|
CCDC105
|
coiled-coil domain containing 105 |
| chr12_+_106751436 | 0.02 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr12_-_46385811 | 0.02 |
ENST00000419565.2
|
SCAF11
|
SR-related CTD-associated factor 11 |
| chr19_-_38714847 | 0.02 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr19_+_10217364 | 0.02 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chrX_+_153237740 | 0.02 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr12_+_52281744 | 0.02 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chr5_+_173416162 | 0.02 |
ENST00000340147.6
|
C5orf47
|
chromosome 5 open reading frame 47 |
| chr1_+_43148625 | 0.02 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr3_-_71802760 | 0.02 |
ENST00000295612.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr17_+_7487146 | 0.02 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr4_+_183370146 | 0.02 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr15_+_90792760 | 0.01 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr22_-_43411106 | 0.01 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr15_+_73735490 | 0.01 |
ENST00000331090.6
ENST00000560581.1 |
C15orf60
|
chromosome 15 open reading frame 60 |
| chr15_+_42697065 | 0.01 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr19_-_18902106 | 0.01 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr10_+_121485588 | 0.01 |
ENST00000361976.2
ENST00000369083.3 |
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr13_-_50018140 | 0.01 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr1_+_43148059 | 0.01 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr1_-_67896095 | 0.00 |
ENST00000370994.4
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr15_+_91498089 | 0.00 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.3 | 0.8 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.8 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0014878 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.3 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.1 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |