Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
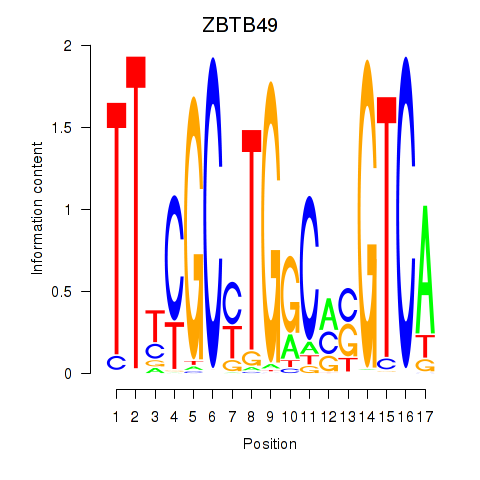
Results for ZBTB49
Z-value: 0.37
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB49 | hg19_v2_chr4_+_4291924_4292011 | -0.29 | 1.5e-01 | Click! |
Activity profile of ZBTB49 motif
Sorted Z-values of ZBTB49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_123676827 | 1.88 |
ENST00000546084.1
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr1_+_79115503 | 1.40 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr16_-_28621353 | 1.18 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr5_-_146833222 | 0.89 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_172428959 | 0.84 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr10_+_90750493 | 0.77 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr11_-_2950642 | 0.73 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chrX_+_30265256 | 0.69 |
ENST00000397548.2
|
MAGEB1
|
melanoma antigen family B, 1 |
| chr10_-_90751038 | 0.67 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chrX_+_9431324 | 0.49 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr19_+_50145328 | 0.44 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr7_-_103629963 | 0.39 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr22_-_42017021 | 0.39 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr8_+_99129513 | 0.39 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr3_-_196242233 | 0.39 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr18_-_11148587 | 0.38 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr11_+_43702322 | 0.35 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr15_+_75639773 | 0.35 |
ENST00000567657.1
|
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr6_-_109777128 | 0.32 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr15_+_93749295 | 0.31 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr19_-_59070239 | 0.29 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr14_+_77924204 | 0.29 |
ENST00000555133.1
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr20_+_33134619 | 0.27 |
ENST00000374837.3
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr3_+_196594727 | 0.27 |
ENST00000445299.2
ENST00000323460.5 ENST00000419026.1 |
SENP5
|
SUMO1/sentrin specific peptidase 5 |
| chr19_-_6481776 | 0.25 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr19_-_54567035 | 0.24 |
ENST00000366170.2
ENST00000425006.2 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr22_+_21996549 | 0.23 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr15_+_90728145 | 0.23 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr8_-_82024290 | 0.23 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr8_+_24151553 | 0.22 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr14_-_74485960 | 0.22 |
ENST00000556242.1
ENST00000334696.6 |
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chrX_+_64887512 | 0.21 |
ENST00000360270.5
|
MSN
|
moesin |
| chrX_-_153210107 | 0.20 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr3_+_52232102 | 0.19 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr7_+_99006232 | 0.19 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr1_+_9294822 | 0.19 |
ENST00000377403.2
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr6_-_49604545 | 0.18 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr19_-_51611623 | 0.18 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr2_+_234160217 | 0.17 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr1_+_240255166 | 0.16 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr5_-_180229833 | 0.16 |
ENST00000307826.4
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr1_+_46806837 | 0.15 |
ENST00000537428.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr2_+_234160340 | 0.15 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr12_+_53835508 | 0.14 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr18_+_32558208 | 0.14 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_-_53525406 | 0.14 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr2_-_118943930 | 0.13 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr2_+_30670077 | 0.13 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr8_+_58907104 | 0.13 |
ENST00000361488.3
|
FAM110B
|
family with sequence similarity 110, member B |
| chr7_-_44613494 | 0.13 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr10_+_94352956 | 0.12 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chrX_-_151903184 | 0.11 |
ENST00000357916.4
ENST00000393869.3 |
MAGEA12
|
melanoma antigen family A, 12 |
| chr12_+_53835383 | 0.11 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr12_+_53835539 | 0.11 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr16_-_18937726 | 0.11 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr15_-_55489097 | 0.10 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr20_+_1115821 | 0.10 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr14_+_79745746 | 0.10 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr16_+_69166418 | 0.10 |
ENST00000314423.7
ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chrX_-_151903101 | 0.09 |
ENST00000393900.3
|
MAGEA12
|
melanoma antigen family A, 12 |
| chr6_-_153452356 | 0.09 |
ENST00000206262.1
|
RGS17
|
regulator of G-protein signaling 17 |
| chr8_+_24151620 | 0.09 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr21_-_26979786 | 0.08 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr11_+_63993738 | 0.08 |
ENST00000441250.2
ENST00000279206.3 |
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr16_-_28415162 | 0.08 |
ENST00000398944.3
ENST00000398943.3 ENST00000380876.4 |
EIF3CL
|
eukaryotic translation initiation factor 3, subunit C-like |
| chr15_-_25684110 | 0.08 |
ENST00000232165.3
|
UBE3A
|
ubiquitin protein ligase E3A |
| chr4_+_128886424 | 0.08 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr9_-_116102530 | 0.07 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chr14_-_35591433 | 0.07 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr6_-_43655511 | 0.07 |
ENST00000372133.3
ENST00000372116.1 ENST00000427312.1 |
MRPS18A
|
mitochondrial ribosomal protein S18A |
| chr22_-_29196546 | 0.07 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr20_-_32700075 | 0.07 |
ENST00000374980.2
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| chr2_+_95691417 | 0.07 |
ENST00000309988.4
|
MAL
|
mal, T-cell differentiation protein |
| chr14_+_79746249 | 0.06 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr2_+_95691445 | 0.06 |
ENST00000353004.3
ENST00000354078.3 ENST00000349807.3 |
MAL
|
mal, T-cell differentiation protein |
| chr14_+_79745682 | 0.06 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr1_+_46806452 | 0.06 |
ENST00000536062.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr22_-_20256054 | 0.05 |
ENST00000043402.7
|
RTN4R
|
reticulon 4 receptor |
| chr16_+_28722809 | 0.05 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr19_-_42894420 | 0.05 |
ENST00000597255.1
ENST00000222032.5 |
CNFN
|
cornifelin |
| chr22_-_20255212 | 0.05 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr8_+_95653302 | 0.04 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr3_-_196756646 | 0.04 |
ENST00000439320.1
ENST00000296351.4 ENST00000296350.5 |
MFI2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr10_+_64893039 | 0.04 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr19_-_4182530 | 0.04 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr2_-_48982708 | 0.04 |
ENST00000428232.1
ENST00000405626.1 ENST00000294954.7 |
LHCGR
|
luteinizing hormone/choriogonadotropin receptor |
| chr22_-_29196030 | 0.03 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr22_+_42017459 | 0.03 |
ENST00000405878.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr17_+_54671047 | 0.03 |
ENST00000332822.4
|
NOG
|
noggin |
| chr19_+_18669809 | 0.03 |
ENST00000602094.1
|
KXD1
|
KxDL motif containing 1 |
| chr1_+_24117627 | 0.03 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr15_+_31508174 | 0.03 |
ENST00000559292.2
ENST00000557928.1 |
RP11-16E12.1
|
RP11-16E12.1 |
| chr10_-_46641003 | 0.02 |
ENST00000395721.2
ENST00000374218.2 ENST00000395725.3 ENST00000374346.3 ENST00000417004.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr1_-_151319283 | 0.02 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr16_+_810728 | 0.01 |
ENST00000563941.1
ENST00000545450.2 ENST00000566549.1 |
MSLN
|
mesothelin |
| chr17_+_80009741 | 0.01 |
ENST00000578552.1
ENST00000320548.4 ENST00000581578.1 ENST00000583885.1 ENST00000583641.1 ENST00000581418.1 ENST00000355130.2 ENST00000306823.6 ENST00000392358.2 |
GPS1
|
G protein pathway suppressor 1 |
| chr17_-_3195876 | 0.01 |
ENST00000323404.1
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1 |
| chr4_+_53525573 | 0.01 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr8_-_59572093 | 0.01 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr16_+_811073 | 0.01 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr17_-_80009650 | 0.01 |
ENST00000310496.4
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_-_97401782 | 0.01 |
ENST00000375326.4
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr11_+_117857063 | 0.00 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr16_+_81069433 | 0.00 |
ENST00000299575.4
|
ATMIN
|
ATM interactor |
| chr17_-_80231557 | 0.00 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB49
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.8 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 0.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 1.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:1903487 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 1.9 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.8 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.0 | 0.0 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.4 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |