Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for ZBTB7A_ZBTB7C
Z-value: 0.70
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
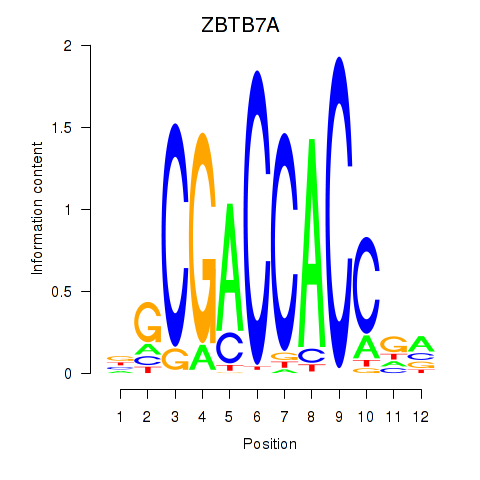
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
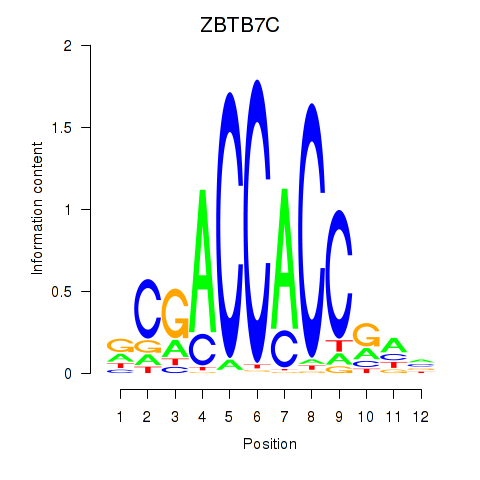
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7C | hg19_v2_chr18_-_45663666_45663732, hg19_v2_chr18_-_45935663_45935793 | -0.56 | 3.5e-03 | Click! |
| ZBTB7A | hg19_v2_chr19_-_4065730_4065909 | -0.34 | 9.8e-02 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_17354587 | 2.27 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr8_+_17354617 | 2.22 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr1_+_169075554 | 1.19 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_-_156721389 | 0.88 |
ENST00000537739.1
|
HDGF
|
hepatoma-derived growth factor |
| chr1_-_156721502 | 0.84 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr10_-_101380121 | 0.83 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr4_-_47916543 | 0.75 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr1_-_38273840 | 0.70 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr17_+_40834580 | 0.69 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr19_+_10197463 | 0.69 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_-_235405168 | 0.64 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr14_+_59655369 | 0.64 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr11_-_117747434 | 0.59 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747607 | 0.58 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_+_101419187 | 0.52 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chrX_-_151143140 | 0.52 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr2_-_74692473 | 0.51 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr1_+_182992545 | 0.49 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr4_+_74718906 | 0.48 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr1_-_156722015 | 0.47 |
ENST00000368209.5
|
HDGF
|
hepatoma-derived growth factor |
| chr7_+_26191809 | 0.46 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr15_-_62352659 | 0.46 |
ENST00000249837.3
|
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr6_+_64282447 | 0.44 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr10_-_105615164 | 0.42 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr9_+_140500087 | 0.41 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr17_-_1531635 | 0.41 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr15_-_64126084 | 0.37 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr1_-_21616901 | 0.37 |
ENST00000436918.2
ENST00000374893.6 |
ECE1
|
endothelin converting enzyme 1 |
| chr8_+_27629459 | 0.35 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr14_+_29236269 | 0.34 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr6_-_82462425 | 0.34 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr1_-_111506562 | 0.34 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr8_-_101963482 | 0.33 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_+_167691191 | 0.33 |
ENST00000392121.3
ENST00000474859.1 |
MPZL1
|
myelin protein zero-like 1 |
| chr9_-_74525847 | 0.32 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr1_+_167691185 | 0.32 |
ENST00000359523.2
|
MPZL1
|
myelin protein zero-like 1 |
| chr6_-_46459675 | 0.32 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chrX_+_153991025 | 0.32 |
ENST00000369550.5
|
DKC1
|
dyskeratosis congenita 1, dyskerin |
| chr4_-_146101304 | 0.32 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr7_+_77167343 | 0.30 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr17_+_79990058 | 0.30 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr22_+_21369316 | 0.30 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr9_+_132934835 | 0.30 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr1_+_15480197 | 0.29 |
ENST00000400796.3
ENST00000434578.2 ENST00000376008.2 |
TMEM51
|
transmembrane protein 51 |
| chr17_+_79989937 | 0.27 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr5_-_132113036 | 0.27 |
ENST00000378706.1
|
SEPT8
|
septin 8 |
| chr12_-_49351148 | 0.27 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr9_-_133814455 | 0.26 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr12_-_56224546 | 0.25 |
ENST00000357606.3
ENST00000547445.1 |
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr17_-_1532106 | 0.25 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr4_-_7069760 | 0.25 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr21_-_47648665 | 0.25 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr15_-_41408339 | 0.25 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr3_-_164914640 | 0.25 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr5_-_132113559 | 0.24 |
ENST00000448933.1
|
SEPT8
|
septin 8 |
| chr8_+_145202939 | 0.24 |
ENST00000423230.2
ENST00000398656.4 |
MROH1
|
maestro heat-like repeat family member 1 |
| chr5_-_2751762 | 0.24 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr6_-_33290580 | 0.24 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr5_-_138739739 | 0.24 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr1_+_38273818 | 0.24 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr5_-_132112907 | 0.24 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr12_-_42631529 | 0.24 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr1_+_38273988 | 0.23 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr7_+_66093851 | 0.23 |
ENST00000275532.3
|
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr5_-_132112921 | 0.22 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr15_+_92396920 | 0.22 |
ENST00000318445.6
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr20_+_37590942 | 0.22 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr2_+_48667983 | 0.21 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr1_+_167905894 | 0.21 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr1_-_95007193 | 0.21 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr19_+_40697514 | 0.21 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr12_+_22199108 | 0.21 |
ENST00000229329.2
|
CMAS
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr17_+_6926339 | 0.21 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr6_+_30689401 | 0.21 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr10_-_99094458 | 0.21 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr12_+_107349497 | 0.21 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr5_+_72861560 | 0.20 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr7_-_94285511 | 0.20 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr3_-_179754556 | 0.20 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr5_-_132113083 | 0.20 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr1_-_92952433 | 0.20 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_94285472 | 0.19 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr9_+_74764278 | 0.19 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr7_+_66093908 | 0.19 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr15_-_41408409 | 0.19 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr19_+_49622646 | 0.18 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr16_-_49315731 | 0.18 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr19_+_45394477 | 0.18 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr9_-_96213828 | 0.18 |
ENST00000423591.1
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr16_+_610407 | 0.18 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chr20_-_61557821 | 0.18 |
ENST00000354665.4
ENST00000370368.1 ENST00000395343.1 ENST00000395340.1 |
DIDO1
|
death inducer-obliterator 1 |
| chr7_-_51384451 | 0.18 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr5_+_76506706 | 0.17 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr8_-_140715294 | 0.17 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
| chr3_-_136471204 | 0.17 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr2_+_149402553 | 0.17 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr8_+_22298578 | 0.17 |
ENST00000240139.5
ENST00000289963.8 ENST00000397775.3 |
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr19_-_59070239 | 0.17 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr13_+_110959598 | 0.17 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr14_-_71276211 | 0.17 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr9_+_109625378 | 0.17 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr5_-_114515734 | 0.16 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr19_+_35634146 | 0.16 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr14_-_24584138 | 0.16 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr17_-_30185971 | 0.16 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr11_-_117747327 | 0.16 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr9_+_74526384 | 0.15 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr1_+_4714792 | 0.15 |
ENST00000378190.3
|
AJAP1
|
adherens junctions associated protein 1 |
| chr6_-_109703600 | 0.15 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr20_-_17662705 | 0.15 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr7_+_77167376 | 0.15 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_-_112430427 | 0.15 |
ENST00000449743.1
ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168
|
transmembrane protein 168 |
| chr8_-_38853990 | 0.15 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chr20_+_36149602 | 0.15 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr8_-_101964231 | 0.15 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr2_+_46769798 | 0.15 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr9_-_131790464 | 0.14 |
ENST00000417224.1
ENST00000416629.1 ENST00000372559.1 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr8_-_134584152 | 0.14 |
ENST00000521180.1
ENST00000517668.1 ENST00000319914.5 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_-_167905225 | 0.14 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr4_-_76598296 | 0.14 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr14_+_29234870 | 0.14 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr5_-_139283982 | 0.14 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr17_+_73201754 | 0.14 |
ENST00000583569.1
ENST00000245544.4 ENST00000579324.1 ENST00000541827.1 ENST00000579298.1 ENST00000447371.2 |
NUP85
|
nucleoporin 85kDa |
| chr12_-_52867569 | 0.14 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr1_+_93811438 | 0.13 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr8_-_101963677 | 0.13 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_42441204 | 0.13 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr16_+_215965 | 0.13 |
ENST00000356815.3
|
HBM
|
hemoglobin, mu |
| chr15_-_75743915 | 0.13 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr19_-_12945362 | 0.13 |
ENST00000590404.1
ENST00000592204.1 |
RTBDN
|
retbindin |
| chr10_+_60145155 | 0.13 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr5_+_140071011 | 0.13 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr19_-_49622348 | 0.13 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr4_+_72053017 | 0.13 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr14_+_33408449 | 0.12 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_111217603 | 0.12 |
ENST00000369769.2
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr7_-_94285402 | 0.12 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr4_+_95373037 | 0.12 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr9_-_139094988 | 0.12 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chrX_+_133507389 | 0.12 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr22_+_42229100 | 0.12 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr17_-_40428359 | 0.12 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr19_-_663277 | 0.12 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr2_+_242255297 | 0.12 |
ENST00000401990.1
ENST00000407971.1 ENST00000436795.1 ENST00000411484.1 ENST00000434955.1 ENST00000402092.2 ENST00000441533.1 ENST00000443492.1 ENST00000437066.1 ENST00000429791.1 |
SEPT2
|
septin 2 |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr1_-_167906277 | 0.12 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr6_+_30689350 | 0.12 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr1_-_22469459 | 0.11 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr12_-_107487604 | 0.11 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr2_+_191513789 | 0.11 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr18_+_55102917 | 0.11 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr15_+_92397051 | 0.11 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr15_-_62352570 | 0.11 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr1_+_45792541 | 0.10 |
ENST00000334815.3
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr2_+_58655461 | 0.10 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr17_+_29158962 | 0.10 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr12_-_56221330 | 0.10 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr8_+_75896731 | 0.10 |
ENST00000262207.4
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr5_-_140070897 | 0.10 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr6_-_109703634 | 0.10 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr19_-_49222956 | 0.10 |
ENST00000599703.1
ENST00000318083.6 ENST00000419611.1 ENST00000377367.3 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr4_-_105416039 | 0.10 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr11_-_1330834 | 0.10 |
ENST00000525159.1
ENST00000317204.6 ENST00000542915.1 ENST00000527938.1 ENST00000530541.1 ENST00000263646.7 |
TOLLIP
|
toll interacting protein |
| chr12_+_132312931 | 0.09 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr5_-_137878887 | 0.09 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr3_+_15468862 | 0.09 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr9_+_135037334 | 0.09 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr1_+_162467595 | 0.09 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr6_-_36953833 | 0.09 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr14_-_102771516 | 0.09 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr1_-_84972248 | 0.09 |
ENST00000370645.4
ENST00000370641.3 |
GNG5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr17_-_58469687 | 0.09 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr19_-_13044494 | 0.09 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr2_+_242255275 | 0.09 |
ENST00000391971.2
|
SEPT2
|
septin 2 |
| chr3_-_113415441 | 0.09 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr1_+_167906056 | 0.08 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr12_-_58240470 | 0.08 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr16_-_2059797 | 0.08 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr8_-_74884511 | 0.08 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr1_+_89246647 | 0.08 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr6_+_73331520 | 0.08 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr8_-_74884482 | 0.08 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr8_-_59572093 | 0.08 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr17_-_40835076 | 0.08 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr11_-_64013663 | 0.08 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_-_48673552 | 0.08 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr1_+_53098862 | 0.08 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chrX_+_152783131 | 0.08 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr11_-_62752455 | 0.07 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_62752162 | 0.07 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr1_+_25664408 | 0.07 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr17_-_63556414 | 0.07 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr6_+_126070726 | 0.07 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr16_+_12995614 | 0.07 |
ENST00000423335.2
|
SHISA9
|
shisa family member 9 |
| chr8_-_74884399 | 0.07 |
ENST00000520210.1
ENST00000602840.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr14_+_70346125 | 0.07 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr17_-_5372271 | 0.07 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr16_+_66400533 | 0.07 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr11_-_62752429 | 0.07 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chrX_-_128977781 | 0.07 |
ENST00000357166.6
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.3 | 0.8 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 1.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 0.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.4 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.2 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.4 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.3 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.2 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.2 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0061183 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) renal vesicle induction(GO:0072034) |
| 0.0 | 0.1 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.6 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 2.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.7 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.4 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 2.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |