Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
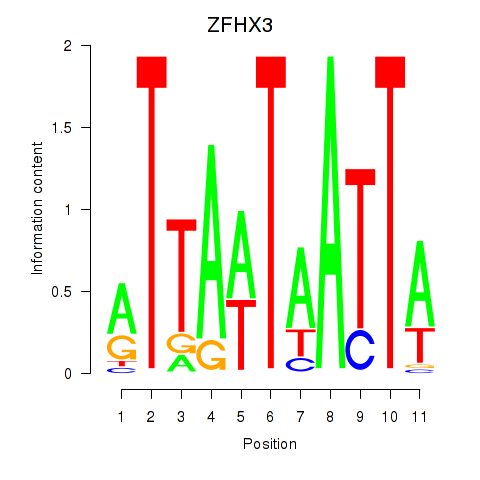
Results for ZFHX3
Z-value: 0.35
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg19_v2_chr16_-_73093597_73093597, hg19_v2_chr16_-_73082274_73082274 | 0.39 | 5.5e-02 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_149365827 | 0.87 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chrX_-_32173579 | 0.33 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr11_+_74459876 | 0.31 |
ENST00000299563.4
|
RNF169
|
ring finger protein 169 |
| chrM_-_14670 | 0.31 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr7_+_100026406 | 0.29 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr5_+_95066823 | 0.28 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr20_-_33735070 | 0.25 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr7_+_130126165 | 0.24 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr22_+_31488433 | 0.24 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr7_+_130126012 | 0.23 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr15_+_76352178 | 0.21 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr12_-_2944184 | 0.20 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr1_+_76540386 | 0.18 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr10_-_75226166 | 0.18 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr12_+_59989791 | 0.18 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_-_110655306 | 0.17 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr11_-_111649015 | 0.17 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_87248975 | 0.16 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr19_-_58864848 | 0.16 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_5410607 | 0.15 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr3_+_46919235 | 0.14 |
ENST00000449590.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr2_+_88047606 | 0.14 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr1_-_168464875 | 0.14 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr12_+_80750134 | 0.14 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chr19_-_36304201 | 0.13 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr11_+_22696314 | 0.13 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr3_-_114477787 | 0.13 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_71868354 | 0.12 |
ENST00000412588.1
|
CALN1
|
calneuron 1 |
| chr22_-_22337204 | 0.12 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr2_-_68052694 | 0.12 |
ENST00000457448.1
|
AC010987.6
|
AC010987.6 |
| chrX_-_18690210 | 0.11 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr20_+_42187608 | 0.11 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr20_+_42187682 | 0.11 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_-_14541060 | 0.10 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr1_+_159557607 | 0.10 |
ENST00000255040.2
|
APCS
|
amyloid P component, serum |
| chr2_+_234621551 | 0.10 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_+_234637754 | 0.10 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chrY_+_20708557 | 0.10 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr6_-_30899924 | 0.10 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr15_+_58724184 | 0.09 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chrY_-_20935572 | 0.09 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr3_+_69915385 | 0.09 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_101659682 | 0.09 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr8_-_145754428 | 0.09 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr12_-_11184006 | 0.08 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr6_+_28317685 | 0.08 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr12_+_131438443 | 0.08 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr12_-_131200810 | 0.08 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr5_+_60933634 | 0.08 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr1_-_247921982 | 0.08 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr3_-_49066811 | 0.08 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr2_+_190541153 | 0.07 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chrM_+_9207 | 0.07 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_+_39703112 | 0.07 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr22_-_30970560 | 0.07 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr7_-_44580861 | 0.07 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr11_-_102576537 | 0.07 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr14_+_39703084 | 0.07 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr2_+_234627424 | 0.07 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chrX_-_15288154 | 0.07 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr18_+_5748793 | 0.06 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chrM_+_8527 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr7_+_150725510 | 0.06 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_-_69111401 | 0.06 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr17_-_39156138 | 0.06 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr6_+_161123270 | 0.06 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_-_29013017 | 0.05 |
ENST00000377175.1
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1 |
| chr6_+_47749718 | 0.05 |
ENST00000489301.2
ENST00000371211.2 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr12_-_68553512 | 0.05 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chrX_+_84258832 | 0.05 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr19_+_926000 | 0.05 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr9_-_95298314 | 0.05 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr12_+_59989918 | 0.05 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_197036364 | 0.04 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr3_+_69812701 | 0.04 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr4_-_100356551 | 0.04 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_-_30881134 | 0.04 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr4_-_100356291 | 0.04 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr2_+_133174147 | 0.04 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chr7_-_100026280 | 0.03 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr6_+_34204642 | 0.03 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr7_-_7575477 | 0.03 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr5_-_125930929 | 0.03 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr7_+_48211048 | 0.03 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr19_-_46234119 | 0.03 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr1_-_160549235 | 0.02 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chrX_+_16668278 | 0.02 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr8_-_18541603 | 0.02 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr19_+_50016610 | 0.02 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_-_15474904 | 0.02 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr17_-_26697304 | 0.02 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr1_-_67266939 | 0.02 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr10_+_118083919 | 0.01 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr6_-_32374900 | 0.01 |
ENST00000374995.3
ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2
|
butyrophilin-like 2 (MHC class II associated) |
| chr3_-_164796269 | 0.01 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr1_+_192544857 | 0.01 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr4_-_155511887 | 0.01 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr11_-_125366089 | 0.00 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_+_50061315 | 0.00 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr6_+_25754927 | 0.00 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0046136 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |