Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
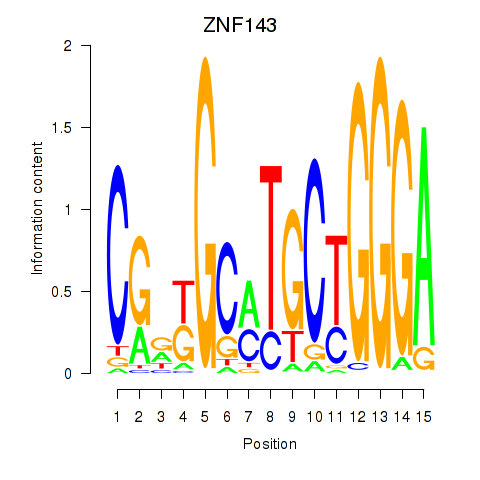
Results for ZNF143
Z-value: 1.11
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.5 | zinc finger protein 143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF143 | hg19_v2_chr11_+_9482512_9482534 | 0.45 | 2.3e-02 | Click! |
Activity profile of ZNF143 motif
Sorted Z-values of ZNF143 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_72385437 | 2.40 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_+_74757050 | 2.19 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr19_+_42746927 | 2.03 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr1_+_244816371 | 2.01 |
ENST00000263831.7
|
DESI2
|
desumoylating isopeptidase 2 |
| chr9_+_35732312 | 1.66 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr20_+_32399093 | 1.59 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr6_+_160148593 | 1.46 |
ENST00000337387.4
|
WTAP
|
Wilms tumor 1 associated protein |
| chr11_-_72492903 | 1.40 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr10_+_124134201 | 1.39 |
ENST00000368990.3
ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr4_+_140222609 | 1.38 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr13_+_25875785 | 1.35 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr18_+_77439775 | 1.34 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr16_+_8891670 | 1.33 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr11_-_72492878 | 1.32 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr17_-_77005860 | 1.29 |
ENST00000591773.1
ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr19_-_44123734 | 1.21 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr12_-_122985067 | 1.19 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr15_+_41913690 | 1.17 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr7_+_112090483 | 1.14 |
ENST00000403825.3
ENST00000429071.1 |
IFRD1
|
interferon-related developmental regulator 1 |
| chr4_-_2965052 | 1.12 |
ENST00000398071.4
ENST00000502735.1 ENST00000314262.6 ENST00000416614.2 |
NOP14
|
NOP14 nucleolar protein |
| chr22_-_50963976 | 1.09 |
ENST00000252785.3
ENST00000395693.3 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr12_-_122985494 | 1.09 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr17_+_26662597 | 1.08 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_+_26662730 | 1.04 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_+_76210267 | 1.04 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr13_-_41706864 | 1.00 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr13_+_25875662 | 0.97 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr11_+_57425209 | 0.96 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr17_-_77005801 | 0.96 |
ENST00000392446.5
|
CANT1
|
calcium activated nucleotidase 1 |
| chr8_+_27632083 | 0.95 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr11_-_3078838 | 0.95 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr19_+_57106647 | 0.95 |
ENST00000328070.6
|
ZNF71
|
zinc finger protein 71 |
| chr1_+_169764163 | 0.91 |
ENST00000413811.2
ENST00000359326.4 ENST00000456684.1 |
C1orf112
|
chromosome 1 open reading frame 112 |
| chr15_-_101835110 | 0.90 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr11_-_3078616 | 0.90 |
ENST00000401769.3
ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS
|
cysteinyl-tRNA synthetase |
| chr19_-_48673580 | 0.89 |
ENST00000427526.2
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr12_+_62654155 | 0.88 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr2_+_26568965 | 0.87 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr16_+_30934376 | 0.85 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr5_-_171433579 | 0.83 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr19_-_10444188 | 0.83 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr22_-_31503490 | 0.82 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr7_+_90032667 | 0.80 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr5_-_171433819 | 0.79 |
ENST00000296933.6
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr19_-_48673552 | 0.79 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr9_+_15422702 | 0.78 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr6_-_116989916 | 0.78 |
ENST00000368576.3
ENST00000368573.1 |
ZUFSP
|
zinc finger with UFM1-specific peptidase domain |
| chr2_-_74757066 | 0.77 |
ENST00000377526.3
|
AUP1
|
ancient ubiquitous protein 1 |
| chr3_+_67048721 | 0.76 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr15_-_101835414 | 0.76 |
ENST00000254193.6
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr20_+_55966444 | 0.75 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr2_-_220252603 | 0.74 |
ENST00000322176.7
ENST00000273075.4 |
DNPEP
|
aspartyl aminopeptidase |
| chr14_+_32546485 | 0.72 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr15_-_50647274 | 0.72 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr6_+_80341000 | 0.72 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr13_+_26828275 | 0.71 |
ENST00000381527.3
|
CDK8
|
cyclin-dependent kinase 8 |
| chr2_+_220143989 | 0.71 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr13_-_50367057 | 0.71 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr2_+_220144052 | 0.71 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr21_+_43919710 | 0.71 |
ENST00000398341.3
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr19_-_2456922 | 0.71 |
ENST00000582871.1
ENST00000325327.3 |
LMNB2
|
lamin B2 |
| chr20_+_46130671 | 0.71 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr7_+_26191809 | 0.70 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr12_+_57881740 | 0.70 |
ENST00000262027.5
ENST00000315473.5 |
MARS
|
methionyl-tRNA synthetase |
| chr15_-_50647370 | 0.69 |
ENST00000558970.1
ENST00000396464.3 ENST00000560825.1 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr2_-_120124258 | 0.69 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr8_+_27632047 | 0.69 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_+_19578033 | 0.69 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr14_-_51135036 | 0.69 |
ENST00000324679.4
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr3_+_94657086 | 0.69 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr17_+_46184911 | 0.67 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr20_+_46130601 | 0.67 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr12_+_22199108 | 0.67 |
ENST00000229329.2
|
CMAS
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr6_+_11094266 | 0.66 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr2_-_120124383 | 0.66 |
ENST00000334816.7
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr1_-_35497283 | 0.66 |
ENST00000373333.1
|
ZMYM6
|
zinc finger, MYM-type 6 |
| chr19_+_532049 | 0.65 |
ENST00000606136.1
|
CDC34
|
cell division cycle 34 |
| chr14_+_32546274 | 0.64 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr8_-_74884511 | 0.63 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr16_+_67880820 | 0.63 |
ENST00000567105.1
|
NUTF2
|
nuclear transport factor 2 |
| chr16_+_3074002 | 0.62 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr17_-_1619535 | 0.62 |
ENST00000573075.1
ENST00000574306.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr3_+_127771212 | 0.61 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr20_+_46130619 | 0.61 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr8_+_141521386 | 0.61 |
ENST00000220913.5
ENST00000519533.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr12_+_26111823 | 0.60 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr17_-_26662440 | 0.60 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr8_-_144897549 | 0.60 |
ENST00000356994.2
ENST00000320476.3 |
SCRIB
|
scribbled planar cell polarity protein |
| chr5_-_32313082 | 0.60 |
ENST00000382142.3
|
MTMR12
|
myotubularin related protein 12 |
| chr17_-_1619491 | 0.59 |
ENST00000570416.1
ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr19_+_44556158 | 0.59 |
ENST00000434772.3
ENST00000585552.1 |
ZNF223
|
zinc finger protein 223 |
| chr18_+_55711575 | 0.59 |
ENST00000356462.6
ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_+_28284803 | 0.58 |
ENST00000586220.1
ENST00000588784.1 ENST00000591549.1 ENST00000585827.1 ENST00000588636.1 ENST00000587188.1 |
CTC-459F4.3
|
CTC-459F4.3 |
| chr8_-_74884482 | 0.58 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr12_+_53662073 | 0.58 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr1_-_53163992 | 0.58 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr12_+_53662110 | 0.57 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr14_-_65569057 | 0.57 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr10_+_60094735 | 0.57 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr14_-_65569244 | 0.56 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr14_-_65569186 | 0.56 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr12_+_77158021 | 0.56 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr3_-_98620500 | 0.56 |
ENST00000326840.6
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr2_+_232063260 | 0.55 |
ENST00000349938.4
|
ARMC9
|
armadillo repeat containing 9 |
| chr19_+_28284357 | 0.55 |
ENST00000588122.1
ENST00000586784.1 ENST00000590628.1 ENST00000592404.1 ENST00000588318.1 ENST00000585917.1 |
CTC-459F4.3
|
CTC-459F4.3 |
| chr8_-_74884399 | 0.55 |
ENST00000520210.1
ENST00000602840.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr19_+_14544099 | 0.55 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr15_+_40453204 | 0.54 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr1_-_2458026 | 0.53 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr17_-_26662464 | 0.53 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr2_+_25015968 | 0.53 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr19_+_58838369 | 0.52 |
ENST00000329665.4
|
ZSCAN22
|
zinc finger and SCAN domain containing 22 |
| chr22_-_42915788 | 0.52 |
ENST00000323013.6
|
RRP7A
|
ribosomal RNA processing 7 homolog A (S. cerevisiae) |
| chr16_+_67881029 | 0.52 |
ENST00000569436.2
ENST00000568396.2 |
NUTF2
|
nuclear transport factor 2 |
| chr5_-_141338627 | 0.52 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr19_-_36705547 | 0.52 |
ENST00000304116.5
|
ZNF565
|
zinc finger protein 565 |
| chr16_-_18937726 | 0.52 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr9_-_131038214 | 0.52 |
ENST00000609374.1
|
GOLGA2
|
golgin A2 |
| chr19_+_44598534 | 0.51 |
ENST00000336976.6
|
ZNF224
|
zinc finger protein 224 |
| chr22_-_38245304 | 0.51 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr2_+_232063436 | 0.51 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr12_+_62654119 | 0.51 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr2_-_202316260 | 0.50 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr8_-_74884459 | 0.50 |
ENST00000522337.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr2_+_97202480 | 0.50 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr2_+_68694678 | 0.50 |
ENST00000303795.4
|
APLF
|
aprataxin and PNKP like factor |
| chr19_+_44488330 | 0.50 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr16_-_69419473 | 0.50 |
ENST00000566750.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr15_-_50647347 | 0.49 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr16_-_68344830 | 0.49 |
ENST00000263997.6
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand |
| chr19_+_14551066 | 0.49 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr18_+_56807096 | 0.49 |
ENST00000588875.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr1_+_156052354 | 0.48 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr17_+_5389605 | 0.48 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr9_-_131038266 | 0.48 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chr8_+_27631903 | 0.47 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_-_73851285 | 0.47 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr2_-_18741882 | 0.47 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr20_-_62339190 | 0.47 |
ENST00000324228.2
ENST00000609142.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chrX_+_48334549 | 0.46 |
ENST00000019019.2
ENST00000348411.2 ENST00000396894.4 |
FTSJ1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr3_-_48229846 | 0.46 |
ENST00000302506.3
ENST00000351231.3 ENST00000437972.1 |
CDC25A
|
cell division cycle 25A |
| chr11_+_66025167 | 0.46 |
ENST00000394067.2
ENST00000316924.5 ENST00000421552.1 ENST00000394078.1 |
KLC2
|
kinesin light chain 2 |
| chr11_+_450255 | 0.46 |
ENST00000308020.5
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr18_-_32924372 | 0.46 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chrX_-_119005735 | 0.46 |
ENST00000371442.2
|
RNF113A
|
ring finger protein 113A |
| chr19_-_3761673 | 0.46 |
ENST00000316757.3
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr1_+_244816237 | 0.45 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr19_+_37862054 | 0.45 |
ENST00000483919.1
ENST00000588911.1 ENST00000436120.2 ENST00000587349.1 |
ZNF527
|
zinc finger protein 527 |
| chr1_+_182992545 | 0.45 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr11_-_113644491 | 0.45 |
ENST00000200135.3
|
ZW10
|
zw10 kinetochore protein |
| chr12_+_53895052 | 0.45 |
ENST00000552857.1
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr2_-_148778258 | 0.45 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr5_-_175815565 | 0.45 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr7_+_2394445 | 0.45 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr5_+_67584174 | 0.44 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_47916613 | 0.44 |
ENST00000381538.3
ENST00000329043.3 |
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr2_-_9695847 | 0.44 |
ENST00000310823.3
ENST00000497134.1 |
ADAM17
|
ADAM metallopeptidase domain 17 |
| chr1_+_45805728 | 0.44 |
ENST00000539779.1
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr1_-_153931052 | 0.44 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr15_+_44829255 | 0.44 |
ENST00000261868.5
ENST00000424492.3 |
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr2_-_239112347 | 0.44 |
ENST00000457149.1
ENST00000254654.3 |
ILKAP
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr1_+_33938236 | 0.44 |
ENST00000361328.3
ENST00000373413.2 |
ZSCAN20
|
zinc finger and SCAN domain containing 20 |
| chr16_-_3074231 | 0.43 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr3_-_122233723 | 0.43 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr2_-_148778323 | 0.43 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr20_-_62339315 | 0.43 |
ENST00000440854.1
ENST00000607873.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chr14_+_32546145 | 0.43 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_-_133747551 | 0.43 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr19_-_47249679 | 0.43 |
ENST00000263280.6
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr11_+_66024737 | 0.42 |
ENST00000531240.1
ENST00000417856.1 |
KLC2
|
kinesin light chain 2 |
| chr1_+_27561104 | 0.42 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr17_+_5390220 | 0.42 |
ENST00000381165.3
|
MIS12
|
MIS12 kinetochore complex component |
| chr5_+_74807581 | 0.42 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr12_+_32832134 | 0.42 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr8_-_74884341 | 0.41 |
ENST00000284811.8
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr18_+_56806701 | 0.41 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chrX_-_118986911 | 0.41 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr18_-_51884204 | 0.41 |
ENST00000577499.1
ENST00000584040.1 ENST00000581310.1 |
STARD6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr4_-_47916543 | 0.41 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr19_+_50145328 | 0.40 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr16_-_12009735 | 0.40 |
ENST00000439887.2
ENST00000434724.2 |
GSPT1
|
G1 to S phase transition 1 |
| chr9_-_34048873 | 0.40 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr1_-_156571254 | 0.40 |
ENST00000438976.2
ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4
|
G patch domain containing 4 |
| chr8_-_21966893 | 0.40 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr16_-_3073933 | 0.40 |
ENST00000574151.1
|
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr16_+_23690138 | 0.40 |
ENST00000300093.4
|
PLK1
|
polo-like kinase 1 |
| chr10_+_120967072 | 0.40 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr17_+_45726803 | 0.40 |
ENST00000535458.2
ENST00000583648.1 |
KPNB1
|
karyopherin (importin) beta 1 |
| chr15_+_44829334 | 0.39 |
ENST00000535391.1
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr5_+_66300446 | 0.39 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_66112555 | 0.39 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr8_-_124054587 | 0.39 |
ENST00000259512.4
|
DERL1
|
derlin 1 |
| chr3_-_50329835 | 0.39 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr1_+_27561007 | 0.39 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr15_-_34502197 | 0.39 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr11_+_2421718 | 0.39 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr20_-_30795511 | 0.38 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr19_+_531713 | 0.38 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr3_+_169490834 | 0.37 |
ENST00000392733.1
|
MYNN
|
myoneurin |
| chr2_-_65357225 | 0.37 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF143
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 1.8 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.6 | 2.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.4 | 1.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.4 | 1.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.4 | 1.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 2.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 1.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 2.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.3 | 1.7 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 1.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.3 | 1.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.3 | 1.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 1.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 | 1.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.3 | 0.8 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 2.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 1.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 0.6 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 0.6 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.2 | 0.6 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.2 | 0.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 0.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 1.0 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0051598 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 1.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.4 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.5 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.0 | GO:0090261 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.3 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.6 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.6 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 2.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.6 | GO:1902947 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.3 | GO:0072233 | thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.2 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 1.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 1.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 1.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.1 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 2.0 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.7 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 1.7 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.9 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.2 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.9 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0071484 | response to high light intensity(GO:0009644) regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.5 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.4 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.4 | 1.5 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 0.9 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.5 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 1.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.5 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.3 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.3 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 2.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0035061 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 3.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.6 | 1.8 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.4 | 2.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 2.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 1.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 1.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.6 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.2 | 1.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.7 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 1.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.0 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 2.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 1.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 2.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.4 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0030346 | potassium:proton antiporter activity(GO:0015386) protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 2.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.7 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 1.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.7 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 2.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 4.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 2.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.5 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |