Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
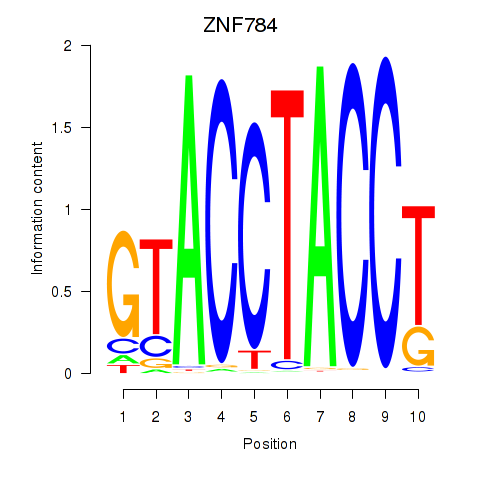
Results for ZNF784
Z-value: 0.58
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg19_v2_chr19_-_56135928_56135967 | 0.24 | 2.4e-01 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_184943610 | 2.21 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr17_+_77020224 | 1.39 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020325 | 1.37 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_+_18284477 | 1.04 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr12_-_52911718 | 0.87 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr17_+_77020146 | 0.78 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_7562334 | 0.73 |
ENST00000593942.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr1_-_1149506 | 0.68 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr21_+_41029235 | 0.66 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr6_-_31550192 | 0.64 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr19_-_42498231 | 0.52 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_-_42498369 | 0.52 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr1_+_90287480 | 0.51 |
ENST00000394593.3
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr4_-_153274078 | 0.50 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_+_52626898 | 0.48 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr3_-_42306248 | 0.45 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr2_-_74692473 | 0.45 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chrX_+_144908928 | 0.43 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr5_-_159827033 | 0.40 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr5_-_159827073 | 0.40 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr1_-_115238207 | 0.39 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr3_+_32993065 | 0.38 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr17_-_1394940 | 0.37 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chr11_+_5009424 | 0.36 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr2_+_47168313 | 0.36 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr16_-_57831914 | 0.36 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr3_-_112218205 | 0.34 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr11_-_134281812 | 0.34 |
ENST00000392580.1
ENST00000312527.4 |
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr1_+_167298281 | 0.34 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr21_-_36421535 | 0.34 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chrX_-_80065146 | 0.33 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr11_+_33061543 | 0.32 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr5_+_112074029 | 0.32 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr21_-_36421626 | 0.32 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr15_+_89346699 | 0.31 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr8_+_38261880 | 0.31 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr13_-_33760216 | 0.31 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr15_+_40453204 | 0.31 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr6_-_161695042 | 0.30 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_+_171283331 | 0.30 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr1_-_186649543 | 0.30 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_+_231191875 | 0.30 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr2_+_47168630 | 0.29 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr6_-_161695074 | 0.29 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_4540486 | 0.29 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_30295036 | 0.28 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr16_-_57831676 | 0.28 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr10_-_76859247 | 0.27 |
ENST00000472493.2
ENST00000605915.1 ENST00000478873.2 |
DUSP13
|
dual specificity phosphatase 13 |
| chr6_+_30294612 | 0.27 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr17_+_19281034 | 0.27 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr3_-_112218378 | 0.27 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr17_+_73717551 | 0.27 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr6_-_100912785 | 0.26 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr12_-_12837423 | 0.26 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr12_-_4754339 | 0.26 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chrX_-_154842538 | 0.25 |
ENST00000369439.4
|
TMLHE
|
trimethyllysine hydroxylase, epsilon |
| chr2_-_145277569 | 0.25 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_158526693 | 0.25 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr2_-_145277640 | 0.24 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_30634297 | 0.24 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr1_+_205473720 | 0.24 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr19_+_39936186 | 0.24 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr4_+_71248795 | 0.24 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr11_+_73358594 | 0.24 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr3_-_195310802 | 0.23 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr17_-_10600818 | 0.23 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chrX_-_49041242 | 0.23 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr19_+_39936267 | 0.23 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr11_+_64008443 | 0.23 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr19_+_39936317 | 0.22 |
ENST00000598725.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr2_+_173940668 | 0.22 |
ENST00000375213.3
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr17_-_19281203 | 0.22 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr5_-_158526756 | 0.22 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr11_+_65154070 | 0.22 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr12_-_49525175 | 0.22 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr16_-_3661578 | 0.22 |
ENST00000294008.3
|
SLX4
|
SLX4 structure-specific endonuclease subunit |
| chr7_+_3340989 | 0.21 |
ENST00000404826.2
ENST00000389531.3 |
SDK1
|
sidekick cell adhesion molecule 1 |
| chr11_+_64008525 | 0.21 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr14_+_105452094 | 0.21 |
ENST00000551606.1
ENST00000547315.1 |
C14orf79
|
chromosome 14 open reading frame 79 |
| chr5_+_169010638 | 0.21 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr17_+_73717516 | 0.21 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr10_-_96122682 | 0.21 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr3_-_48130314 | 0.20 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr1_-_207095212 | 0.20 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_+_90032667 | 0.20 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr7_+_45067265 | 0.20 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chrX_-_23761317 | 0.20 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr12_+_122242597 | 0.20 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr15_+_89346657 | 0.20 |
ENST00000439576.2
|
ACAN
|
aggrecan |
| chr6_-_45983581 | 0.19 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr12_+_115800817 | 0.19 |
ENST00000547948.1
|
RP11-116D17.1
|
HCG2038717; Uncharacterized protein |
| chr6_-_45983549 | 0.19 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr1_+_50575292 | 0.19 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_48130707 | 0.18 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr7_-_150652924 | 0.18 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr3_-_147124547 | 0.18 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr4_+_71263599 | 0.18 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr11_-_19262486 | 0.18 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr15_+_71839566 | 0.18 |
ENST00000357769.4
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr9_+_130565487 | 0.18 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr1_+_33219592 | 0.18 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr4_+_39046615 | 0.17 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr7_+_128577972 | 0.17 |
ENST00000357234.5
ENST00000477535.1 ENST00000479582.1 ENST00000464557.1 ENST00000402030.2 |
IRF5
|
interferon regulatory factor 5 |
| chr8_+_11561660 | 0.17 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr9_-_116037840 | 0.17 |
ENST00000374206.3
|
CDC26
|
cell division cycle 26 |
| chr3_-_121379739 | 0.17 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_54653313 | 0.17 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr5_-_178772424 | 0.16 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr3_+_51863433 | 0.16 |
ENST00000444293.1
|
IQCF3
|
IQ motif containing F3 |
| chr19_+_49622646 | 0.15 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr6_+_123100853 | 0.15 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr11_+_74862032 | 0.15 |
ENST00000289575.5
ENST00000341411.4 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr9_-_99381660 | 0.15 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr11_-_36619771 | 0.15 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr11_-_8290263 | 0.15 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chrX_-_106243451 | 0.15 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chrX_-_54384425 | 0.15 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr4_+_158141899 | 0.15 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr1_-_207095324 | 0.14 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr9_-_130693048 | 0.14 |
ENST00000388747.4
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr16_-_67190152 | 0.14 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr20_+_44036620 | 0.14 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_+_158141843 | 0.14 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158141806 | 0.14 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr5_-_1524015 | 0.14 |
ENST00000283415.3
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr1_-_25256368 | 0.14 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr3_-_9834375 | 0.14 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr11_+_65122216 | 0.14 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr9_+_116037922 | 0.14 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr2_-_24583583 | 0.14 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr17_-_56494713 | 0.14 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr1_+_110026544 | 0.14 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr1_-_161208013 | 0.14 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_64216748 | 0.13 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr20_+_20348740 | 0.13 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr2_-_127864839 | 0.13 |
ENST00000409400.1
ENST00000357970.3 ENST00000393040.3 ENST00000348750.4 ENST00000259238.4 ENST00000346226.3 ENST00000393041.3 ENST00000351659.3 ENST00000352848.3 ENST00000316724.5 |
BIN1
|
bridging integrator 1 |
| chr7_+_119913688 | 0.13 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr6_-_43496605 | 0.13 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr5_-_137674000 | 0.13 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr17_+_73717407 | 0.13 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr4_+_158142750 | 0.13 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr8_+_80523321 | 0.13 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr22_+_20067738 | 0.13 |
ENST00000351989.3
ENST00000383024.2 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chr13_-_96329048 | 0.13 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr5_+_137225158 | 0.13 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr9_-_99382065 | 0.13 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr21_-_43916433 | 0.12 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr1_+_24645807 | 0.12 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_187476721 | 0.12 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr17_-_56494882 | 0.12 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr9_-_112260531 | 0.12 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr11_+_125439298 | 0.12 |
ENST00000278903.6
ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24
|
etoposide induced 2.4 |
| chr18_-_5419797 | 0.12 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr11_+_76494253 | 0.12 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr1_-_161207986 | 0.12 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_50574585 | 0.12 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_142636440 | 0.11 |
ENST00000458732.1
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr21_-_43916296 | 0.11 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr3_+_50712672 | 0.11 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr7_-_103629963 | 0.11 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr11_-_10590118 | 0.11 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr19_+_13858593 | 0.11 |
ENST00000221554.8
|
CCDC130
|
coiled-coil domain containing 130 |
| chr4_+_113568207 | 0.11 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_210111570 | 0.11 |
ENST00000367019.1
ENST00000472886.1 |
SYT14
|
synaptotagmin XIV |
| chr11_-_115375107 | 0.11 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr12_+_53895364 | 0.11 |
ENST00000552817.1
ENST00000394357.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr19_+_38042240 | 0.11 |
ENST00000592533.1
|
ZNF540
|
zinc finger protein 540 |
| chr19_-_375970 | 0.11 |
ENST00000346878.2
|
THEG
|
theg spermatid protein |
| chrX_+_70798261 | 0.11 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr3_+_85008089 | 0.11 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr3_-_42623428 | 0.11 |
ENST00000423701.2
ENST00000420163.1 ENST00000416880.1 ENST00000536332.1 ENST00000264454.3 ENST00000273156.7 |
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr1_-_25291475 | 0.10 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr19_-_376011 | 0.10 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr7_+_26677490 | 0.10 |
ENST00000409974.3
|
C7orf71
|
chromosome 7 open reading frame 71 |
| chrY_+_16636354 | 0.10 |
ENST00000339174.5
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr1_-_160616804 | 0.10 |
ENST00000538290.1
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr2_-_220025548 | 0.10 |
ENST00000356853.5
|
NHEJ1
|
nonhomologous end-joining factor 1 |
| chr1_+_228327943 | 0.10 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr14_-_55658323 | 0.10 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr17_-_74707037 | 0.10 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr13_+_24153488 | 0.10 |
ENST00000382258.4
ENST00000382263.3 |
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_24645865 | 0.10 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24646002 | 0.10 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_56494908 | 0.10 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr22_-_37640277 | 0.09 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr6_-_47010061 | 0.09 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr3_-_51994694 | 0.09 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr9_-_110251836 | 0.09 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr17_-_10276319 | 0.09 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr7_+_100199800 | 0.09 |
ENST00000223061.5
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr11_+_45944190 | 0.09 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr20_+_44036900 | 0.09 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_-_55658252 | 0.09 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chrX_-_55024967 | 0.09 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_-_92951607 | 0.09 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_130418888 | 0.09 |
ENST00000310992.4
|
KLF14
|
Kruppel-like factor 14 |
| chr1_+_228327923 | 0.09 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr18_+_905104 | 0.09 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr1_+_173837488 | 0.09 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr22_+_21321531 | 0.09 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.5 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.2 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.1 | 0.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.3 | GO:0033121 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.6 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0090179 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0006127 | glycerol-3-phosphate metabolic process(GO:0006072) glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 2.1 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.4 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 1.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.6 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.2 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0033647 | host cell cytoplasm(GO:0030430) host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) host cell cytoplasm part(GO:0033655) |
| 0.0 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 1.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 3.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |