Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
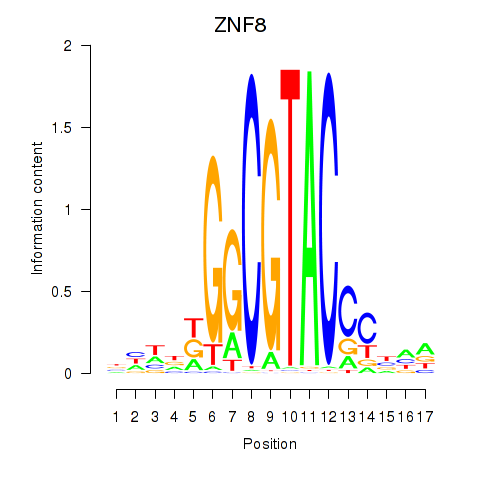
Results for ZNF8
Z-value: 0.33
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | 0.15 | 4.6e-01 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_79258444 | 0.83 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_+_79258547 | 0.81 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr11_+_13690249 | 0.67 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr6_+_53659746 | 0.62 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr2_+_173600514 | 0.42 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173600565 | 0.41 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_186344802 | 0.37 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr19_+_37998031 | 0.36 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr6_-_31763408 | 0.32 |
ENST00000444930.2
|
VARS
|
valyl-tRNA synthetase |
| chr2_+_169926047 | 0.31 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_+_138677515 | 0.27 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr4_+_170541678 | 0.25 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr4_+_170541835 | 0.24 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr17_+_34136459 | 0.24 |
ENST00000588240.1
ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr5_-_93447333 | 0.21 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr1_-_246670519 | 0.21 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr17_+_75181292 | 0.20 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr13_+_45563721 | 0.20 |
ENST00000361121.2
|
GPALPP1
|
GPALPP motifs containing 1 |
| chr1_+_214161272 | 0.20 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chrX_+_102631248 | 0.19 |
ENST00000361298.4
ENST00000372645.3 ENST00000372635.1 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr5_+_10353780 | 0.18 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr4_+_170541660 | 0.17 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr14_+_105941118 | 0.16 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr22_-_37415475 | 0.15 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr2_-_240322643 | 0.13 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr2_+_87144738 | 0.13 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr4_+_186347388 | 0.12 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr11_+_46402583 | 0.12 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_+_6615241 | 0.11 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr6_+_106959718 | 0.11 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr4_-_52904425 | 0.10 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr22_+_30163340 | 0.10 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr19_-_18392422 | 0.10 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr17_+_48624450 | 0.10 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr1_+_70671363 | 0.10 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr7_+_1748798 | 0.09 |
ENST00000561626.1
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr6_-_26056695 | 0.08 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr1_-_70671216 | 0.08 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr10_+_135192695 | 0.08 |
ENST00000368539.4
ENST00000278060.5 ENST00000357296.3 |
PAOX
|
polyamine oxidase (exo-N4-amino) |
| chr3_-_46759314 | 0.08 |
ENST00000315170.7
|
PRSS50
|
protease, serine, 50 |
| chr16_+_29973351 | 0.08 |
ENST00000602948.1
ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219
|
transmembrane protein 219 |
| chr12_-_74686314 | 0.07 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr6_-_116601044 | 0.07 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr1_-_235292250 | 0.07 |
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr11_+_46402482 | 0.06 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_+_37415776 | 0.06 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr10_-_13043697 | 0.06 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr11_+_13690200 | 0.05 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr1_-_55266926 | 0.05 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr9_+_96026230 | 0.05 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr22_+_37415676 | 0.04 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr1_+_186344883 | 0.04 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr4_-_186347099 | 0.04 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr5_+_102455968 | 0.04 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr7_-_74489609 | 0.04 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr16_+_21964662 | 0.04 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr22_+_37415728 | 0.04 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr19_+_35739782 | 0.04 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739897 | 0.04 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr22_+_37415700 | 0.04 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr1_+_186344945 | 0.03 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr10_+_1034646 | 0.03 |
ENST00000360059.5
ENST00000545048.1 |
GTPBP4
|
GTP binding protein 4 |
| chr2_+_172290707 | 0.03 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr9_-_32552551 | 0.02 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr10_+_1034338 | 0.02 |
ENST00000360803.4
ENST00000538293.1 |
GTPBP4
|
GTP binding protein 4 |
| chr1_-_100715372 | 0.02 |
ENST00000370131.3
ENST00000370132.4 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr2_+_198365122 | 0.01 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr16_-_53537105 | 0.00 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr13_+_21750780 | 0.00 |
ENST00000309594.4
|
MRP63
|
mitochondrial ribosomal protein 63 |
| chr13_-_21750659 | 0.00 |
ENST00000400018.3
ENST00000314759.5 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr22_-_30162924 | 0.00 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.7 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.2 | GO:0061114 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.0 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |