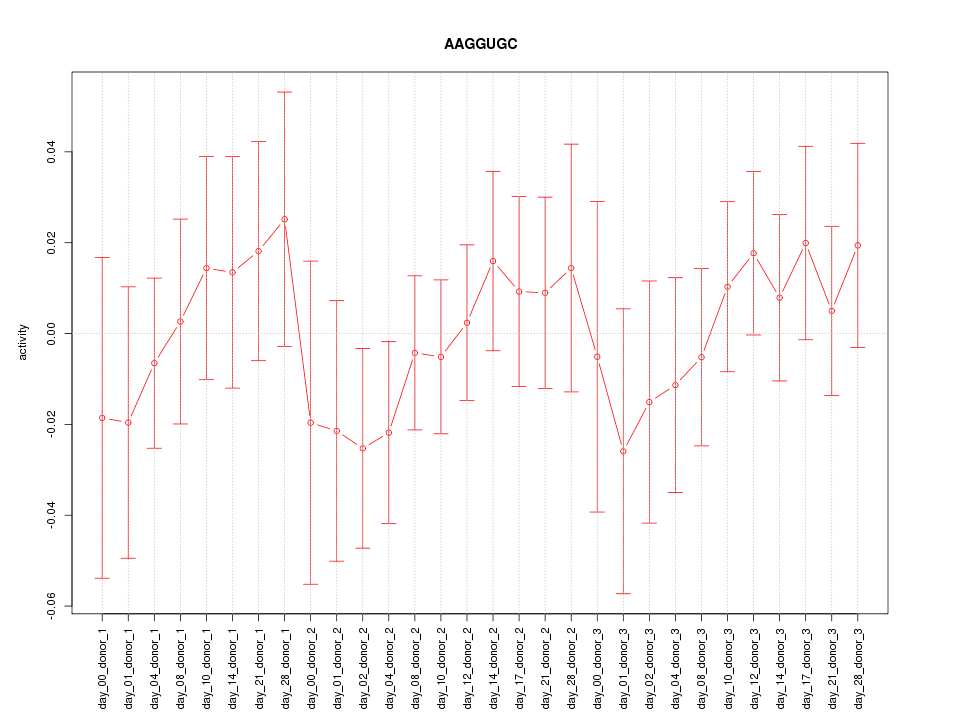
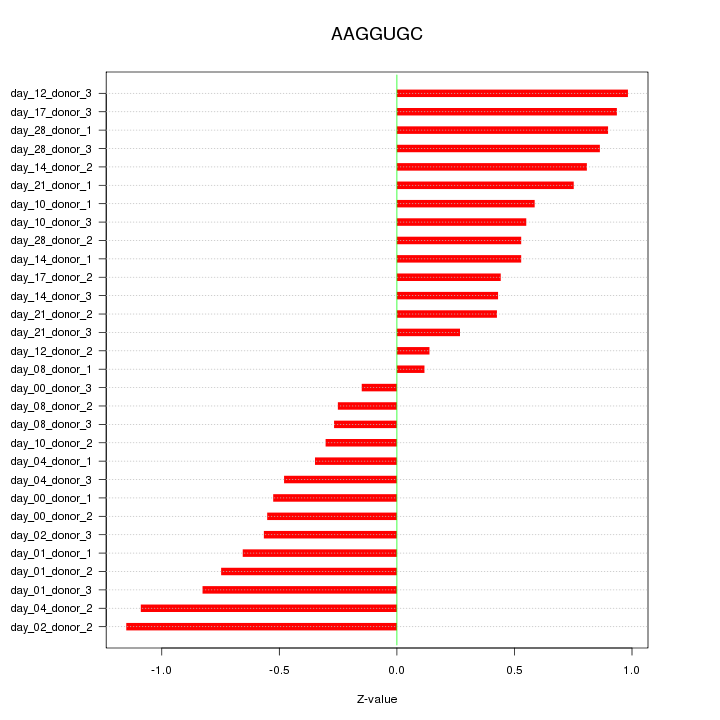
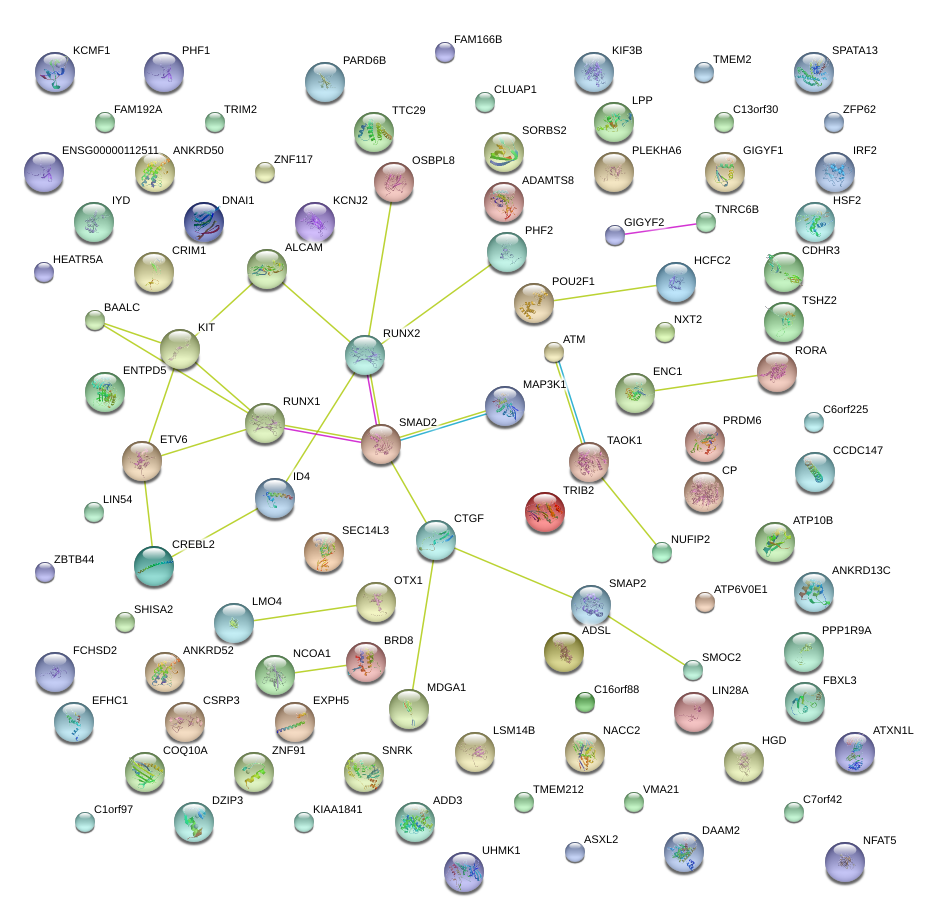
|
chr6_-_132272311
|
3.110
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr6_+_112408673
|
2.558
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr13_+_43355685
|
2.497
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr4_+_55523906
|
2.132
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr6_+_39760148
|
1.443
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr6_+_39760772
|
1.313
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr15_-_61521478
|
1.203
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_-_108464344
|
1.178
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr15_-_60884615
|
1.116
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_+_12764755
|
0.944
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr15_-_60919642
|
0.941
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_+_104458235
|
0.920
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr9_-_35563895
|
0.850
|
NM_001099951
NM_001164310
|
FAM166B
|
family with sequence similarity 166, member B
|
|
chr2_+_63277191
|
0.829
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr5_-_73937248
|
0.781
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr2_+_63277936
|
0.765
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_+_87794150
|
0.764
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr9_-_74383301
|
0.675
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr9_+_96338671
|
0.617
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr11_-_130184459
|
0.607
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr1_+_40839368
|
0.603
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr2_+_12856813
|
0.596
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr21_-_36421586
|
0.594
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_204329043
|
0.584
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr1_+_40859016
|
0.558
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr2_+_24807345
|
0.546
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr22_-_30867944
|
0.514
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr13_-_77601296
|
0.504
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr13_-_26625164
|
0.475
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr9_+_34458810
|
0.463
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr1_+_40840304
|
0.460
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr1_+_40862471
|
0.459
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr5_+_56110899
|
0.459
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr1_+_162467594
|
0.452
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr6_+_19837599
|
0.451
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr4_-_147866963
|
0.449
|
NM_031956
|
TTC29
|
tetratricopeptide repeat domain 29
|
|
chr16_+_69599868
|
0.437
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr12_-_56652118
|
0.436
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr17_-_27621163
|
0.433
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr1_+_162466963
|
0.408
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr5_+_122424840
|
0.402
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr8_+_104152876
|
0.401
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr3_+_108308336
|
0.394
|
NM_014648
|
DZIP3
|
DAZ interacting protein 3, zinc finger
|
|
chr6_+_52285804
|
0.383
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr3_+_187930720
|
0.376
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_105603656
|
0.366
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr3_+_187871473
|
0.361
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_+_68165660
|
0.355
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr20_+_30865435
|
0.350
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr18_-_45456925
|
0.346
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr19_-_23578226
|
0.329
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr18_-_45457454
|
0.328
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr6_+_52284948
|
0.327
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr11_-_72852957
|
0.324
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr13_+_24734788
|
0.324
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr4_+_154125589
|
0.314
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr6_+_122720685
|
0.311
|
NM_001135564
NM_001243094
NM_004506
|
HSF2
|
heat shock transcription factor 2
|
|
chr22_+_40440664
|
0.310
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr12_+_11802726
|
0.302
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr16_+_71879893
|
0.301
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr4_-_125633716
|
0.292
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr10_+_111767685
|
0.290
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr4_+_154074269
|
0.289
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_+_233561997
|
0.286
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr9_-_138987096
|
0.285
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr11_+_108093558
|
0.285
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr7_+_66386129
|
0.283
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr4_-_186697065
|
0.283
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_150690027
|
0.282
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr20_+_60697471
|
0.274
|
NM_144703
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae)
|
|
chr2_+_85198176
|
0.270
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr6_-_119031230
|
0.269
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr1_-_70820363
|
0.259
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr10_+_106113513
|
0.258
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr7_+_94536926
|
0.256
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr5_-_180288275
|
0.246
|
NM_001172638
NM_152283
|
ZFP62
|
zinc finger protein 62 homolog (mouse)
|
|
chr4_-_186578122
|
0.245
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186877869
|
0.245
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_+_150565655
|
0.243
|
NM_001017980
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr2_-_26101301
|
0.239
|
NM_018263
|
ASXL2
|
additional sex combs like 2 (Drosophila)
|
|
chr4_-_83931973
|
0.239
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr16_+_3550916
|
0.235
|
NM_015041
|
CLUAP1
|
clusterin associated protein 1
|
|
chr17_+_27717942
|
0.233
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr2_+_36583369
|
0.231
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr7_+_94539209
|
0.231
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr16_+_3559893
|
0.230
|
NM_024793
|
CLUAP1
|
clusterin associated protein 1
|
|
chr1_+_26737232
|
0.225
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr2_+_61292999
|
0.222
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr7_-_100286869
|
0.221
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr16_-_57219951
|
0.220
|
NM_024946
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr3_+_105085553
|
0.219
|
NM_001243280
NM_001243281
NM_001243283
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr3_-_120401401
|
0.218
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr20_+_49348050
|
0.217
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr6_-_37665631
|
0.215
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr4_-_185395616
|
0.203
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr3_+_43328001
|
0.199
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr3_-_148939784
|
0.195
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr5_-_160279045
|
0.195
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr14_-_31889736
|
0.195
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr7_-_64451413
|
0.191
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr12_+_56661026
|
0.191
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr20_+_51588945
|
0.190
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_+_167189948
|
0.190
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr3_+_171561138
|
0.190
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chrX_+_108780235
|
0.186
|
NM_001242618
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr5_-_58334936
|
0.185
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_56660548
|
0.183
|
NM_144576
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr1_+_167298280
|
0.178
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_+_108779009
|
0.176
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr12_+_69864065
|
0.176
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr20_+_51801821
|
0.174
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr2_-_202316227
|
0.168
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr1_+_172502012
|
0.166
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr6_-_16761600
|
0.163
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr3_-_169899519
|
0.162
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr5_-_59783885
|
0.162
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_40834571
|
0.161
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr4_-_83933961
|
0.160
|
NM_001115007
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr15_+_73344824
|
0.157
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr5_+_140514787
|
0.155
|
NM_015669
|
PCDHB5
|
protocadherin beta 5
|
|
chr14_-_95623665
|
0.153
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr21_-_36260972
|
0.153
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr20_+_278200
|
0.152
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chrX_+_108780058
|
0.149
|
NM_001242617
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr1_+_172502291
|
0.148
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr11_-_69519105
|
0.147
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr17_-_7382943
|
0.146
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr4_+_108814419
|
0.145
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_-_55459406
|
0.143
|
NM_152385
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr1_-_23521221
|
0.142
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr4_+_108745718
|
0.139
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chrX_-_24045302
|
0.138
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr8_-_145701717
|
0.137
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr2_-_55459698
|
0.137
|
NM_001135598
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chrX_-_108868250
|
0.137
|
NM_012282
|
KCNE1L
|
KCNE1-like
|
|
chr7_+_75831194
|
0.136
|
NM_001110199
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chrX_-_19688991
|
0.136
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr20_-_4982133
|
0.134
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr16_-_66730466
|
0.133
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr11_-_79151694
|
0.132
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr19_-_22018940
|
0.130
|
NM_003423
|
ZNF43
|
zinc finger protein 43
|
|
chr10_+_111765725
|
0.128
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr7_+_90893735
|
0.127
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr18_+_55888750
|
0.124
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_+_22217369
|
0.124
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr12_-_15374290
|
0.123
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr4_-_186732047
|
0.123
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_-_64211111
|
0.122
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr7_+_21467572
|
0.121
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr6_-_86352635
|
0.121
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_180601121
|
0.120
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr3_+_107241782
|
0.119
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr19_-_7293876
|
0.119
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chrX_-_19905665
|
0.118
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr6_-_24358279
|
0.117
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr6_-_107435635
|
0.117
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr1_-_213189087
|
0.115
|
NM_144567
|
ANGEL2
|
angel homolog 2 (Drosophila)
|
|
chr9_+_125703111
|
0.113
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr11_+_109964086
|
0.111
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr1_-_55681024
|
0.110
|
NM_015306
|
USP24
|
ubiquitin specific peptidase 24
|
|
chr1_-_89530890
|
0.109
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr5_+_140625146
|
0.107
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chrX_+_77166178
|
0.107
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_+_141488314
|
0.107
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr8_+_144731829
|
0.106
|
NM_014789
|
ZNF623
|
zinc finger protein 623
|
|
chr3_-_15900928
|
0.106
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr16_+_28996146
|
0.105
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr6_+_26368405
|
0.105
|
NM_001197246
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr19_+_58341661
|
0.105
|
NM_001204818
|
LOC100293516
|
zinc finger protein
|
|
chr6_+_47624222
|
0.104
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr5_+_31799030
|
0.104
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr7_-_152133071
|
0.104
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr21_-_18985224
|
0.104
|
NM_001130914
NM_006806
|
BTG3
|
BTG family, member 3
|
|
chr2_+_205410465
|
0.103
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr1_-_109506016
|
0.102
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr6_-_86352564
|
0.101
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_-_24383519
|
0.100
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr6_+_80340971
|
0.100
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr6_+_138188352
|
0.100
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_27886443
|
0.100
|
NM_014860
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like
|
|
chr19_-_20748540
|
0.099
|
NM_001159293
|
ZNF737
|
zinc finger protein 737
|
|
chr5_+_153825497
|
0.097
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr1_+_227751218
|
0.095
|
NM_178549
|
ZNF678
|
zinc finger protein 678
|
|
chr10_+_38299525
|
0.095
|
NM_006954
NM_006974
|
ZNF33A
|
zinc finger protein 33A
|
|
chr1_-_202777545
|
0.095
|
NM_006618
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr2_-_96874569
|
0.094
|
NM_020151
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr4_+_157997181
|
0.094
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr11_+_118307072
|
0.093
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr3_-_13114547
|
0.092
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr19_-_19774501
|
0.092
|
NM_020410
|
ATP13A1
|
ATPase type 13A1
|
|
chr1_-_247242111
|
0.092
|
NM_001204220
NM_033213
|
ZNF670-ZNF695
ZNF670
|
ZNF670-ZNF695 readthrough
zinc finger protein 670
|
|
chrX_-_19817907
|
0.092
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_145438437
|
0.091
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr4_-_186732208
|
0.091
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr16_+_28996369
|
0.090
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr1_+_70820485
|
0.090
|
NM_001031693
NM_001036645
NM_001036646
|
HHLA3
|
HERV-H LTR-associating 3
|
|
chr17_+_26698986
|
0.089
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr18_+_55711388
|
0.089
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr20_-_1165116
|
0.088
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr9_+_120466453
|
0.088
|
NM_138554
|
TLR4
|
toll-like receptor 4
|