|
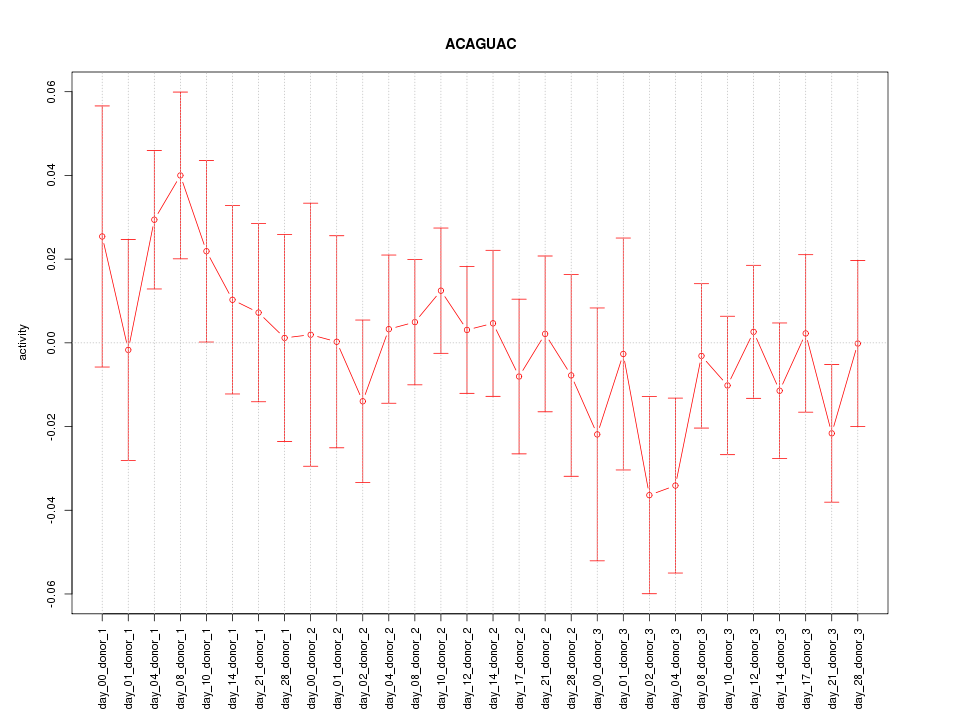
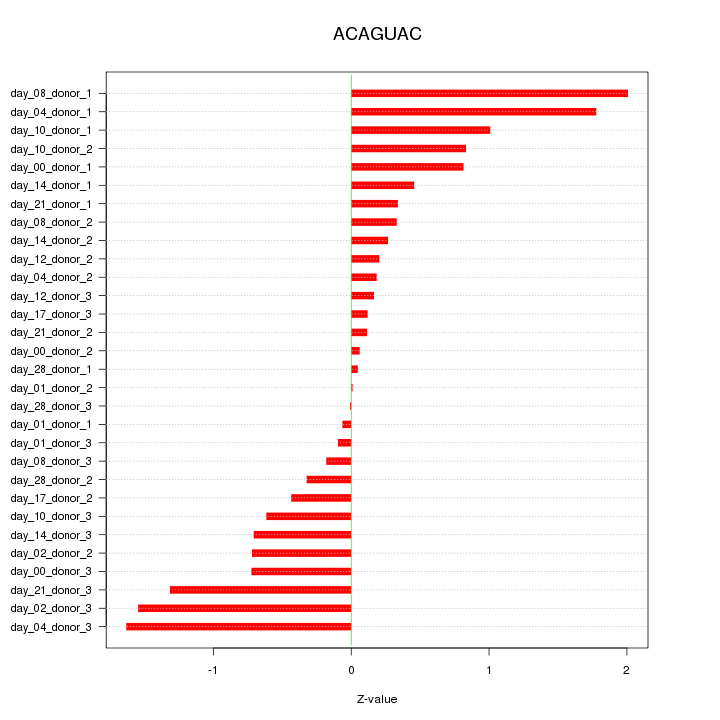
chr6_+_163149001
|
1.475
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr6_+_163148163
|
1.454
|
NM_001080378
NM_152410
|
PACRG
|
PARK2 co-regulated
|
|
chr8_-_23712297
|
1.443
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_+_3841079
|
1.333
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chrY_+_15016018
|
1.318
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr10_+_115803805
|
1.298
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr7_-_148580600
|
0.891
|
NM_001203249
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chrY_+_15016698
|
0.861
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr11_-_115375065
|
0.858
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_-_186649421
|
0.818
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr7_-_148581403
|
0.815
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_-_204459445
|
0.806
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr5_+_149151502
|
0.796
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr6_+_151815174
|
0.791
|
NM_025059
|
C6orf97
|
chromosome 6 open reading frame 97
|
|
chr5_+_149109814
|
0.745
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr20_+_11898536
|
0.739
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr8_+_75896707
|
0.695
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr2_-_217560247
|
0.693
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr2_-_99771186
|
0.657
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr2_-_99758034
|
0.642
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr12_+_56414688
|
0.631
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr8_+_40010986
|
0.618
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr11_-_74660165
|
0.555
|
NM_182969
|
XRRA1
|
X-ray radiation resistance associated 1
|
|
chr2_+_220094478
|
0.553
|
NM_018089
NM_001042410
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1
|
|
chr7_+_73442426
|
0.544
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr6_+_143929316
|
0.540
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr3_+_88188261
|
0.537
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr6_+_143999058
|
0.535
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr10_+_76586299
|
0.532
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr14_+_45431333
|
0.532
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr19_+_10543110
|
0.526
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr6_+_135502445
|
0.518
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_+_244214552
|
0.506
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr13_-_61989654
|
0.503
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr17_+_60556247
|
0.494
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr2_+_191273012
|
0.492
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr20_+_11871364
|
0.489
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr1_+_244216506
|
0.466
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr6_-_111804413
|
0.453
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr10_-_94050799
|
0.450
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr8_-_72268720
|
0.443
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr19_+_10563581
|
0.442
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr6_+_107811272
|
0.438
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr7_-_44887697
|
0.434
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr9_-_14398981
|
0.425
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr5_+_127419407
|
0.421
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr5_-_59783885
|
0.418
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_93963597
|
0.418
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr5_+_153825497
|
0.417
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr4_+_72052354
|
0.416
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr10_+_97889467
|
0.408
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr14_-_61190458
|
0.404
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr1_+_183605207
|
0.400
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr9_-_14314036
|
0.396
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr3_+_23986735
|
0.392
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr21_+_35445808
|
0.388
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr4_+_106816593
|
0.385
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr8_-_56685869
|
0.383
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr5_-_58571916
|
0.382
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_72274466
|
0.373
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_94003002
|
0.372
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr4_-_141075232
|
0.369
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr6_-_10838712
|
0.367
|
NM_001242385
NM_001242957
NM_005906
|
MAK
|
male germ cell-associated kinase
|
|
chr1_-_47779651
|
0.365
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr1_-_169455099
|
0.360
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr5_-_59189620
|
0.360
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_23987514
|
0.358
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr16_+_14164879
|
0.357
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr19_+_10541334
|
0.354
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr22_+_35937351
|
0.351
|
NM_014310
|
RASD2
|
RASD family, member 2
|
|
chr12_+_111843743
|
0.347
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr15_+_33603162
|
0.346
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr1_+_179923870
|
0.346
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr6_+_33387846
|
0.343
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr1_-_173962050
|
0.343
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr4_+_72204769
|
0.341
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr18_-_53255734
|
0.341
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr20_+_61427804
|
0.337
|
NM_018270
|
C20orf20
|
chromosome 20 open reading frame 20
|
|
chr11_-_70507911
|
0.334
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr1_+_220701547
|
0.332
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr6_-_52926588
|
0.331
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr14_-_65438794
|
0.329
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr19_-_33793429
|
0.326
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr22_+_47022657
|
0.325
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr2_+_74056042
|
0.321
|
NM_201647
|
STAMBP
|
STAM binding protein
|
|
chr1_-_86043932
|
0.317
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_-_112726143
|
0.313
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr19_+_10531128
|
0.305
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr11_-_95657255
|
0.305
|
NM_001243571
NM_016156
NM_201278
NM_201281
|
MTMR2
|
myotubularin related protein 2
|
|
chr3_-_183273407
|
0.296
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr1_-_197169635
|
0.292
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr2_+_74056095
|
0.288
|
NM_006463
NM_213622
|
STAMBP
|
STAM binding protein
|
|
chr10_+_120789227
|
0.282
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chrX_+_106449861
|
0.282
|
NM_001169154
NM_173494
|
CXorf41
|
chromosome X open reading frame 41
|
|
chr20_+_51801821
|
0.282
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr15_-_51914770
|
0.279
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr15_-_37391596
|
0.277
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr15_-_59225800
|
0.277
|
NM_001013843
NM_024755
|
SLTM
|
SAFB-like, transcription modulator
|
|
chr5_-_58295758
|
0.276
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_37390372
|
0.275
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr17_-_63557615
|
0.275
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr20_+_51588945
|
0.275
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr11_-_70935807
|
0.274
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr1_-_85930733
|
0.265
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr14_+_75898836
|
0.262
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr2_+_46926049
|
0.261
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr8_-_74659082
|
0.261
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr20_+_58515408
|
0.261
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr16_+_58059281
|
0.261
|
NM_002428
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr10_-_105614952
|
0.260
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr7_+_100026412
|
0.260
|
NM_001194990
NM_001194991
NM_001194992
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr5_+_137688270
|
0.260
|
NM_016604
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr3_+_143692166
|
0.252
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr4_+_15004297
|
0.251
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr4_-_18023358
|
0.246
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr14_+_75894919
|
0.246
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr8_+_110346549
|
0.242
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr12_-_76478737
|
0.237
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr16_-_1429612
|
0.236
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr7_+_8008416
|
0.235
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr8_+_17354562
|
0.235
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chrX_-_19988381
|
0.235
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr17_-_65241305
|
0.234
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr7_+_120590816
|
0.231
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chrX_-_128657453
|
0.231
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr15_-_37393364
|
0.229
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_205473683
|
0.225
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr14_-_57735614
|
0.222
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr5_-_40798263
|
0.222
|
NM_006251
NM_206907
|
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit
|
|
chr16_-_74808699
|
0.219
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr15_-_37392358
|
0.219
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr7_+_100027253
|
0.218
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr14_+_75894508
|
0.216
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr2_-_20527120
|
0.215
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr4_+_38665587
|
0.213
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr2_+_198380294
|
0.213
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr6_-_82462374
|
0.213
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr5_+_141488314
|
0.212
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr3_-_69101236
|
0.211
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr6_+_19837599
|
0.209
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chrX_-_108976509
|
0.209
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr19_-_31840118
|
0.209
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr3_-_15839674
|
0.208
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr9_+_101867401
|
0.207
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr5_-_58652671
|
0.207
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_38495778
|
0.206
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr11_+_121322848
|
0.205
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr6_-_33168369
|
0.205
|
NM_021976
|
RXRB
|
retinoid X receptor, beta
|
|
chr2_+_46926323
|
0.202
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr20_+_58508818
|
0.201
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr21_+_18885104
|
0.201
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr12_+_19592311
|
0.200
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr1_+_82266081
|
0.200
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr2_-_24149899
|
0.200
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr6_+_138483024
|
0.200
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr6_-_13711643
|
0.200
|
NM_005493
|
RANBP9
|
RAN binding protein 9
|
|
chr5_-_114880536
|
0.199
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr15_+_100173182
|
0.198
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr2_+_198381021
|
0.197
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr16_+_67596309
|
0.193
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr8_+_136469557
|
0.193
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr2_-_68479481
|
0.192
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr8_+_17396285
|
0.192
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr8_+_79578258
|
0.191
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr6_-_35464716
|
0.190
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr16_+_71879893
|
0.189
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr18_-_812268
|
0.189
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr16_+_19125243
|
0.189
|
NM_001034841
|
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2
|
|
chr15_-_60884615
|
0.188
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr21_-_27543134
|
0.186
|
NM_000484
NM_001136129
NM_001136130
NM_001204301
NM_001204302
NM_001204303
NM_201413
NM_201414
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr8_-_124553448
|
0.185
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr7_-_44924946
|
0.185
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr7_-_47621741
|
0.184
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr10_-_43903220
|
0.183
|
NM_001098206
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr4_-_78740508
|
0.180
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr3_-_125094003
|
0.180
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr16_-_10674443
|
0.176
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr5_-_115910406
|
0.172
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chrX_+_123095555
|
0.172
|
NM_001042750
NM_001042751
NM_006603
|
STAG2
|
stromal antigen 2
|
|
chr8_-_42698447
|
0.172
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr19_+_2164119
|
0.171
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr11_+_109964086
|
0.171
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr5_-_59064437
|
0.170
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_109065576
|
0.170
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr11_+_11862955
|
0.169
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr1_+_154193294
|
0.168
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr3_-_15900928
|
0.168
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr4_+_106067754
|
0.168
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr15_-_60919642
|
0.167
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr6_+_4021500
|
0.167
|
NM_003913
|
PRPF4B
|
PRP4 pre-mRNA processing factor 4 homolog B (yeast)
|
|
chr4_-_125633716
|
0.166
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr11_-_34535302
|
0.165
|
NM_001243080
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr20_-_52199635
|
0.163
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr15_+_77223942
|
0.163
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr8_-_82024277
|
0.162
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr8_-_124544376
|
0.161
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr4_-_36245893
|
0.160
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr18_+_9137560
|
0.159
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr6_-_166075520
|
0.158
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr13_-_79233266
|
0.157
|
NM_024546
|
RNF219
|
ring finger protein 219
|