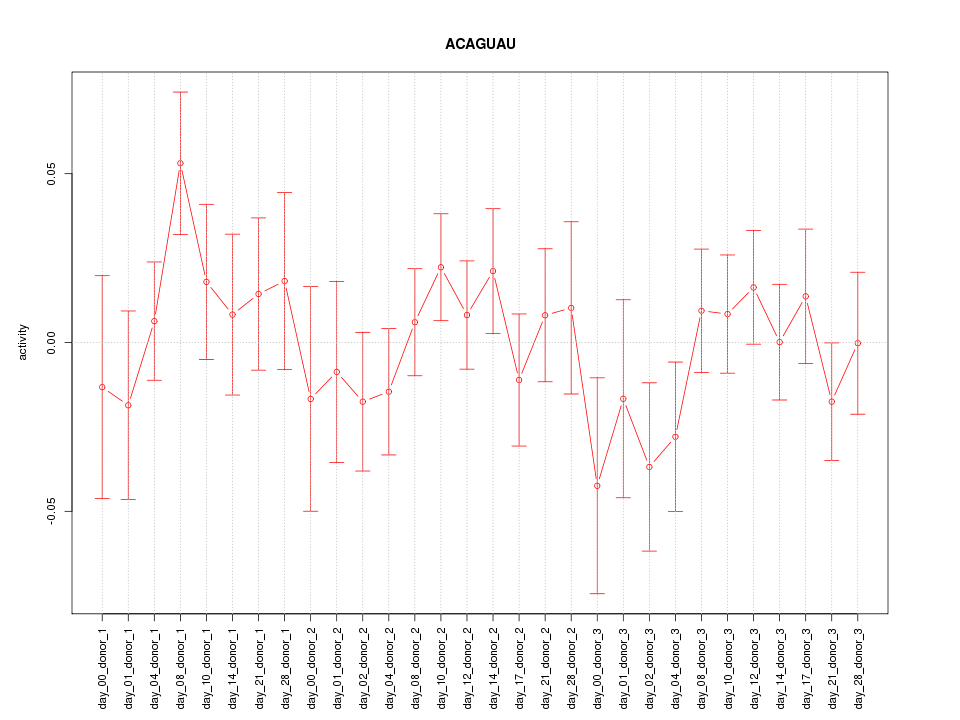
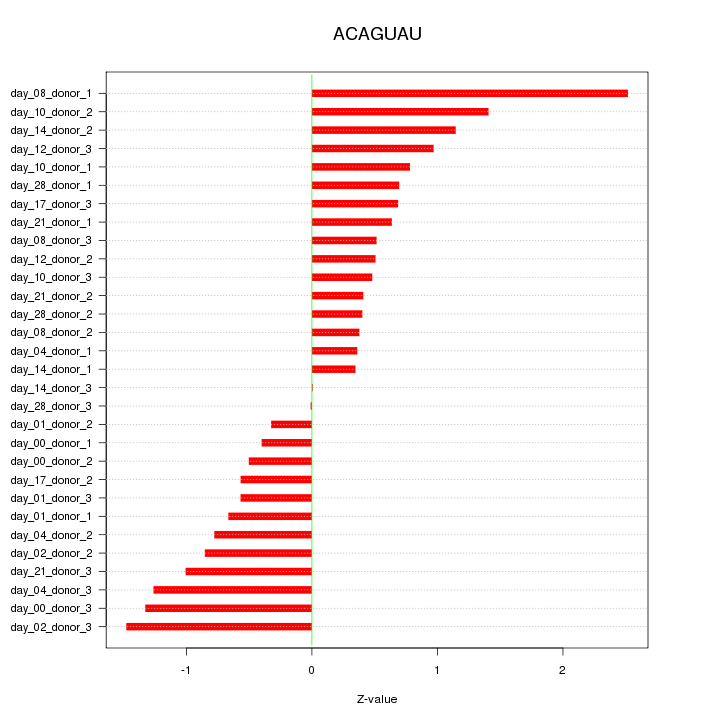
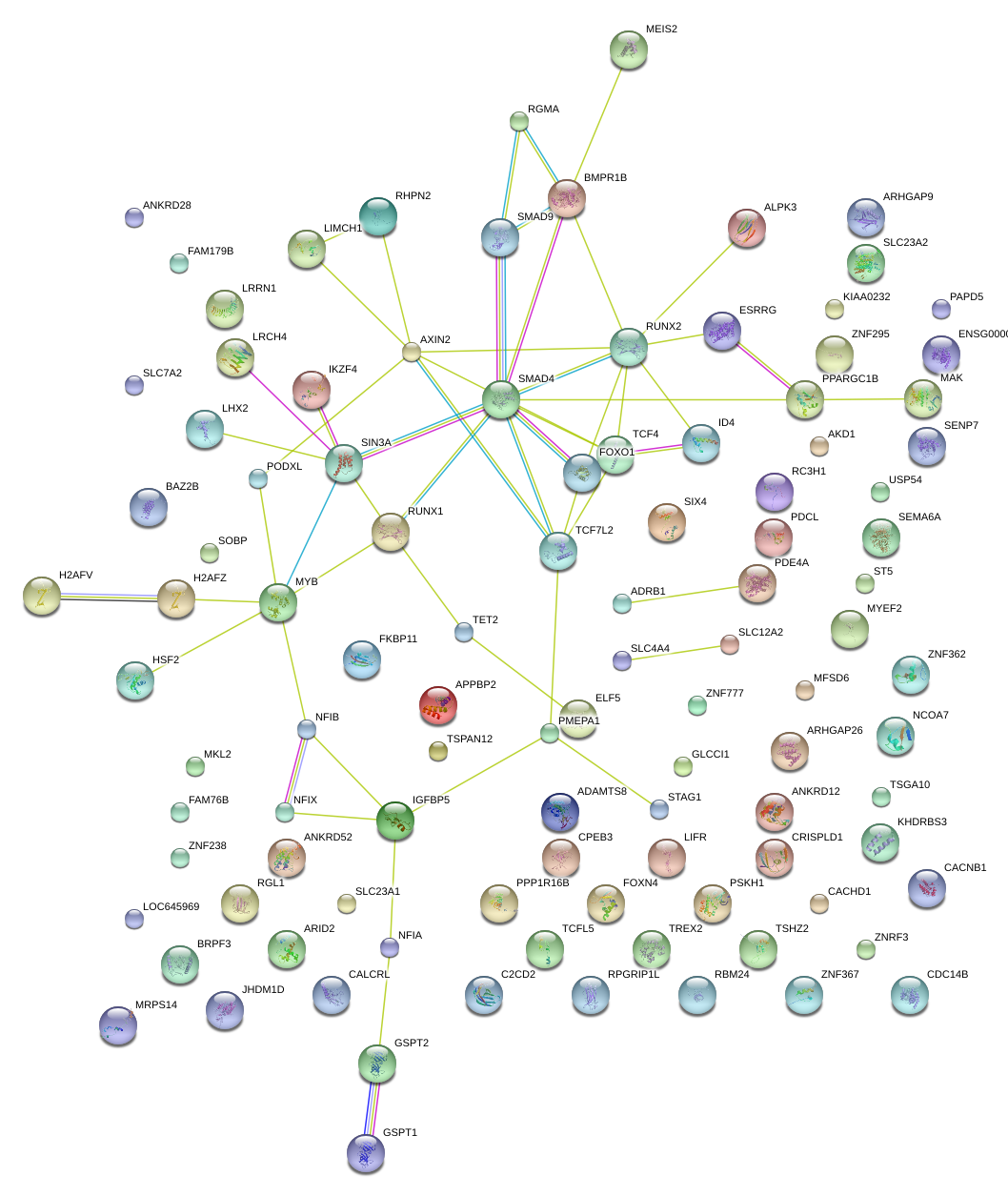
|
chr7_-_131241321
|
2.940
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr15_-_48470453
|
1.840
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr2_-_99771186
|
1.477
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr2_-_99758034
|
1.462
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr8_+_136469557
|
1.317
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr22_+_29279573
|
1.286
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr6_+_126111782
|
1.281
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr16_-_53737713
|
1.264
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr6_+_126240369
|
1.229
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr10_-_75335415
|
1.221
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr3_+_3841079
|
1.219
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr6_+_107811272
|
1.214
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr8_+_75896707
|
1.204
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr6_+_19837599
|
1.203
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr6_+_17281773
|
1.188
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282931
|
1.165
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
1.162
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr1_+_183605207
|
1.155
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr1_+_244214552
|
1.155
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr17_-_63557615
|
1.137
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr19_-_33555757
|
1.128
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr19_+_10543110
|
1.059
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr5_-_38595506
|
1.031
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr1_+_244216506
|
1.021
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr22_+_29313376
|
1.010
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr5_+_127419407
|
0.993
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr4_+_72052354
|
0.972
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_-_115910406
|
0.969
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr6_+_135502445
|
0.969
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr5_-_38556682
|
0.922
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr6_+_126102278
|
0.914
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr20_+_37434294
|
0.913
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr15_-_93632161
|
0.906
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616891
|
0.898
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr19_+_10563581
|
0.880
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr4_+_6784453
|
0.876
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr15_-_93617091
|
0.874
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr6_-_10838712
|
0.870
|
NM_001242385
NM_001242957
NM_005906
|
MAK
|
male germ cell-associated kinase
|
|
chr10_+_115803805
|
0.842
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr20_+_51588945
|
0.838
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr16_+_14164879
|
0.833
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr15_-_93631751
|
0.830
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr20_+_51801821
|
0.827
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr5_+_149109814
|
0.815
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_217262946
|
0.814
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr21_-_43373938
|
0.813
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_+_149151502
|
0.809
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr15_-_93616358
|
0.802
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr19_+_10541334
|
0.789
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr1_+_33722158
|
0.772
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr19_+_10531128
|
0.762
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr4_+_95679075
|
0.761
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr1_+_61547625
|
0.756
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_217311095
|
0.750
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chrX_+_51486479
|
0.747
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr1_+_61542928
|
0.731
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_173962050
|
0.729
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr1_-_216896640
|
0.727
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr15_-_75743776
|
0.717
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr15_-_75744029
|
0.705
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr6_+_36164528
|
0.695
|
NM_015695
|
BRPF3
|
bromodomain and PHD finger containing, 3
|
|
chr15_-_75748123
|
0.694
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr6_+_122720685
|
0.684
|
NM_001135564
NM_001243094
NM_004506
|
HSF2
|
heat shock transcription factor 2
|
|
chr9_-_99329092
|
0.678
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr16_+_67927170
|
0.662
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr8_+_17354562
|
0.652
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr4_+_72204769
|
0.646
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_64936475
|
0.646
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr4_+_41614911
|
0.634
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr12_+_46123495
|
0.633
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr7_-_120498128
|
0.629
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr12_-_56652118
|
0.610
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr14_-_61190458
|
0.609
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr1_+_61547388
|
0.607
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr11_-_95522945
|
0.606
|
NM_144664
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr16_-_12010518
|
0.604
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr7_-_139876718
|
0.597
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr10_-_94050799
|
0.591
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr12_+_56414688
|
0.591
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr13_-_37494370
|
0.570
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr2_-_217560247
|
0.569
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr11_-_34535302
|
0.559
|
NM_001243080
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr4_+_41362750
|
0.558
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_106067754
|
0.557
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr2_-_188312872
|
0.555
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr15_-_37392358
|
0.540
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr9_+_126773851
|
0.534
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr9_-_125590809
|
0.533
|
NM_005388
|
PDCL
|
phosducin-like
|
|
chr7_-_149157866
|
0.531
|
NM_015694
|
ZNF777
|
zinc finger protein 777
|
|
chr11_-_34533117
|
0.530
|
NM_001243081
NM_198381
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr15_-_37391596
|
0.529
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr15_-_37390372
|
0.526
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr8_+_17396285
|
0.525
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr12_-_109747024
|
0.524
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr3_-_15900928
|
0.513
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr13_-_41240607
|
0.512
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr3_-_101231973
|
0.508
|
NM_001077203
NM_020654
|
SENP7
|
SUMO1/sentrin specific peptidase 7
|
|
chr11_-_8832881
|
0.507
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr11_-_8832189
|
0.503
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr14_+_45431333
|
0.501
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr18_+_9137560
|
0.496
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr2_+_191273012
|
0.487
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr3_+_150321065
|
0.486
|
NM_016275
|
SELT
|
selenoprotein T
|
|
chr18_+_48556483
|
0.482
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr2_-_160472960
|
0.471
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr11_-_8932497
|
0.465
|
NM_005418
|
ST5
|
suppression of tumorigenicity 5
|
|
chr16_+_50186809
|
0.462
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr5_+_142150291
|
0.461
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr20_-_4982133
|
0.461
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr17_-_58603492
|
0.460
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr21_-_43430448
|
0.460
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr15_-_37393364
|
0.458
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr3_-_15839674
|
0.456
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_174992552
|
0.456
|
NM_022100
|
MRPS14
|
mitochondrial ribosomal protein S14
|
|
chr20_-_61492945
|
0.456
|
NM_006602
|
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix)
|
|
chr10_-_94003002
|
0.451
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_99382073
|
0.450
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr7_+_8008416
|
0.449
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr21_-_36421586
|
0.449
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_136471186
|
0.447
|
NM_005862
|
STAG1
|
stromal antigen 1
|
|
chr7_-_44887697
|
0.442
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr18_-_53255734
|
0.441
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr10_+_76586299
|
0.436
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr16_-_87525317
|
0.432
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr10_+_120789227
|
0.431
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr5_+_153825497
|
0.423
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr5_-_176981537
|
0.413
|
NM_001190946
|
FAM193B
|
family with sequence similarity 193, member B
|
|
chr17_+_7608413
|
0.411
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr4_+_106816593
|
0.410
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr16_-_68482373
|
0.406
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr6_+_143929316
|
0.406
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr6_-_134496006
|
0.405
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_143999058
|
0.404
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_-_245027808
|
0.403
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr6_-_134497069
|
0.399
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_69812961
|
0.399
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_-_61521478
|
0.397
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_+_79578258
|
0.394
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr12_-_68726036
|
0.388
|
NM_001205028
NM_001205029
NM_017440
NM_020128
|
MDM1
|
Mdm1 nuclear protein homolog (mouse)
|
|
chr15_-_60884615
|
0.380
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr21_-_43346780
|
0.374
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr7_+_73442426
|
0.371
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr6_-_90121966
|
0.370
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr5_-_59783885
|
0.370
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_-_107435635
|
0.369
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr15_+_96869156
|
0.367
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_+_23986735
|
0.361
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr4_-_141075232
|
0.356
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr14_+_73525187
|
0.356
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr3_+_23987514
|
0.352
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr15_+_96876568
|
0.349
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_169455099
|
0.345
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr6_-_134639195
|
0.344
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr16_-_12009804
|
0.344
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr8_-_124553448
|
0.340
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr10_-_61900659
|
0.337
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_134498973
|
0.337
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_134181369
|
0.336
|
NM_152409
|
C5orf24
|
chromosome 5 open reading frame 24
|
|
chr1_-_47779651
|
0.335
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr19_-_47735858
|
0.331
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr15_-_77363519
|
0.327
|
NM_001168412
NM_005724
NM_198902
|
TSPAN3
|
tetraspanin 3
|
|
chr3_+_69915374
|
0.325
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_-_42698447
|
0.325
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr10_-_62149487
|
0.324
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrX_-_108976509
|
0.321
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr8_+_61429390
|
0.321
|
NM_001242644
NM_002865
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr15_-_60919642
|
0.318
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_69812620
|
0.314
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr6_-_139695349
|
0.313
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr19_-_47734215
|
0.313
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr10_-_13390189
|
0.310
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr21_+_44394634
|
0.309
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr7_-_47621741
|
0.309
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr18_+_9136742
|
0.306
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr6_+_33387846
|
0.306
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr3_+_69788562
|
0.305
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_100106132
|
0.305
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr9_-_98270321
|
0.304
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr6_-_139695784
|
0.300
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr1_-_235490801
|
0.300
|
NM_001206794
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr4_+_144434555
|
0.299
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr1_+_26798901
|
0.294
|
NM_005517
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr11_+_109964086
|
0.293
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr11_-_70507911
|
0.291
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr1_+_179923870
|
0.290
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr12_-_102224509
|
0.290
|
NM_024312
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits
|
|
chr10_-_62332666
|
0.289
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr12_+_68042493
|
0.286
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr6_+_148663728
|
0.283
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr10_-_98346793
|
0.282
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_-_52926588
|
0.276
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr3_+_88188261
|
0.276
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr14_+_103800492
|
0.276
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr19_-_33793429
|
0.274
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr9_-_98269686
|
0.274
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr1_+_204485461
|
0.274
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr15_+_96875793
|
0.272
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_-_24149899
|
0.271
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr10_-_62493282
|
0.271
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr20_-_4990913
|
0.269
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|