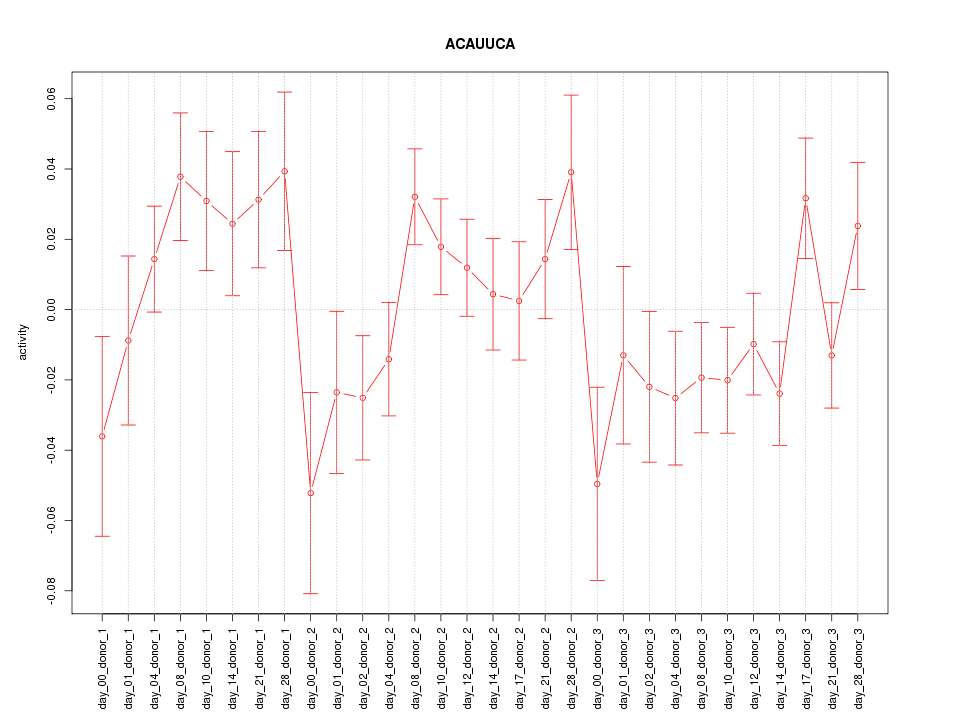
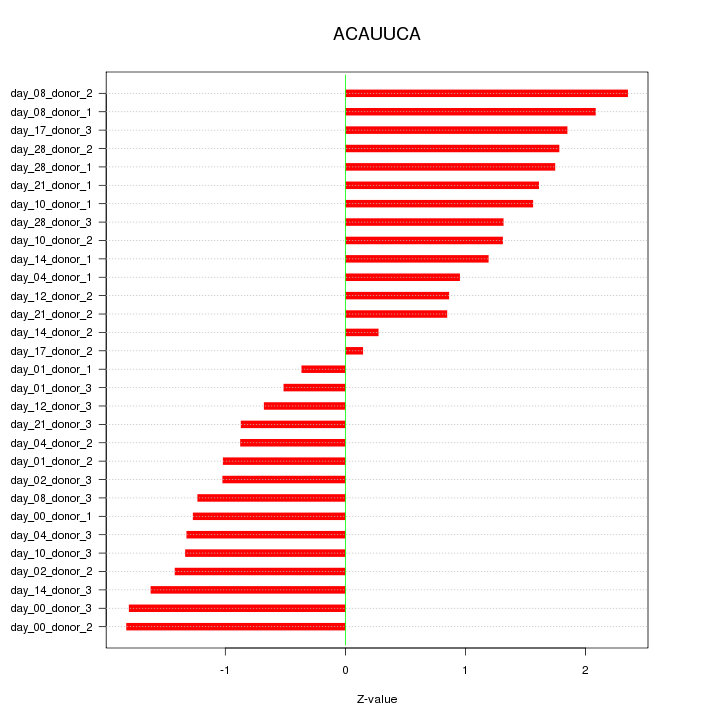
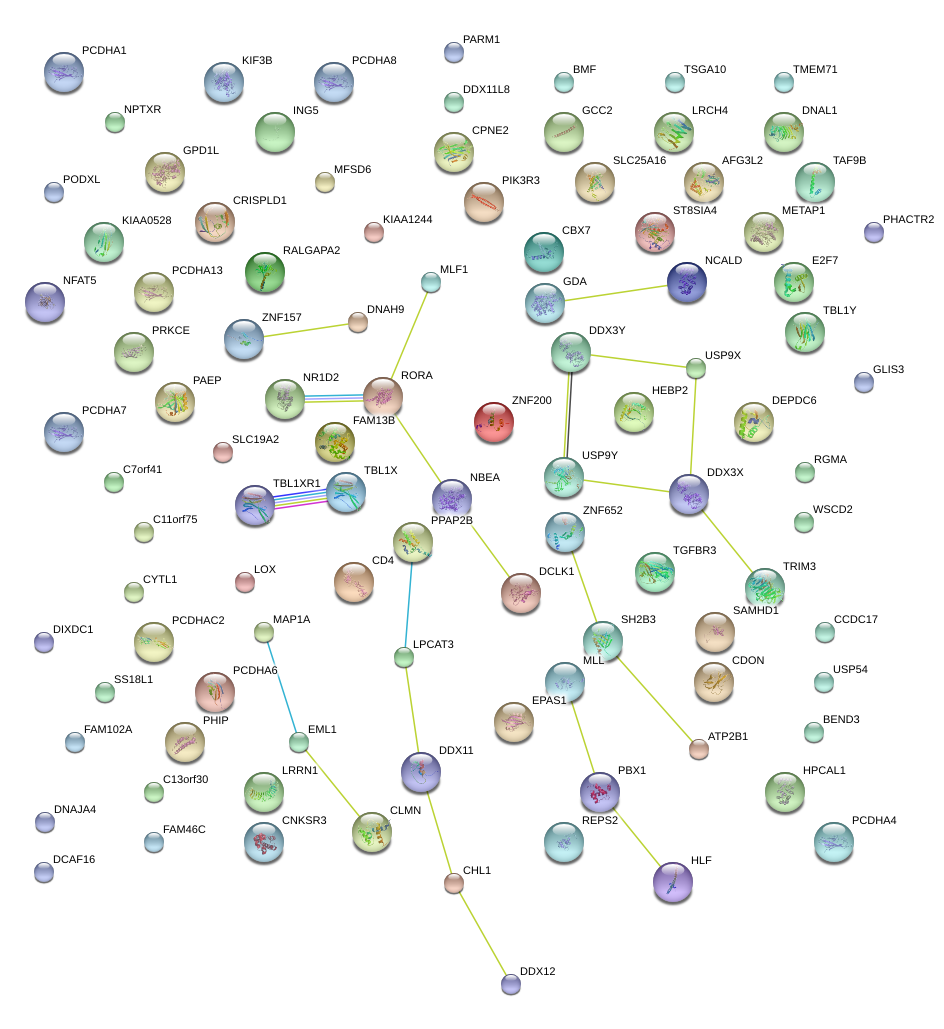
|
chr13_+_43355685
|
7.052
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr7_-_131241321
|
5.533
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr3_+_158288952
|
3.646
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr15_+_43809805
|
3.637
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr1_-_46089638
|
3.615
|
NM_001114938
NM_001190182
|
CCDC17
|
coiled-coil domain containing 17
|
|
chr9_-_4300028
|
3.436
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr9_-_4152182
|
3.359
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr8_+_75896707
|
3.121
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr1_+_118148506
|
3.101
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr22_-_39548448
|
2.904
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr14_-_95786095
|
2.746
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr13_+_36050885
|
2.646
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr4_+_75858205
|
2.605
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr5_-_137368778
|
2.580
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr5_+_140186658
|
2.575
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_+_140254930
|
2.550
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140213968
|
2.541
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr5_+_140220811
|
2.513
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chrY_+_15016018
|
2.460
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr1_-_46598250
|
2.448
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr15_-_93632161
|
2.327
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr13_+_35516390
|
2.230
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr2_-_99771186
|
2.226
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr15_-_93616891
|
2.226
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr2_-_99758034
|
2.198
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr15_-_93617091
|
2.151
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chrX_+_9431314
|
2.100
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr15_+_78556901
|
2.062
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_+_78558522
|
2.059
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_-_93631751
|
2.045
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr11_-_93276513
|
2.038
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chrY_+_14813159
|
2.037
|
NM_004654
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked
|
|
chr15_-_93616358
|
2.032
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr12_-_22697439
|
1.989
|
NM_014802
|
KIAA0528
|
KIAA0528
|
|
chr5_-_100238870
|
1.987
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr5_+_140247767
|
1.986
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr8_-_103136486
|
1.955
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr8_-_133772808
|
1.921
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr15_+_78556485
|
1.918
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr20_-_35580186
|
1.905
|
NM_015474
|
SAMHD1
|
SAM domain and HD domain 1
|
|
chr5_-_121412805
|
1.905
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr5_-_121414011
|
1.876
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr8_-_103137134
|
1.873
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr3_+_3841079
|
1.843
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr14_+_74111577
|
1.764
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr6_-_20212628
|
1.759
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr8_-_102803438
|
1.757
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr14_+_100259445
|
1.738
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chrX_+_9432980
|
1.733
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr16_-_3285359
|
1.723
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chrY_+_15016698
|
1.664
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr12_-_90049818
|
1.660
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr5_+_140165875
|
1.648
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr20_-_20693122
|
1.606
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr13_-_36429798
|
1.584
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr18_-_12377248
|
1.528
|
NM_006796
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr15_-_40398286
|
1.528
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr15_-_40398573
|
1.501
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr17_-_47439326
|
1.480
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr12_+_6898605
|
1.476
|
NM_000616
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr12_-_77459198
|
1.470
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr8_+_120885899
|
1.458
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr20_+_60718473
|
1.447
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr3_+_23986735
|
1.436
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chrX_-_77395108
|
1.435
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr5_+_140207562
|
1.411
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr17_+_53342320
|
1.406
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr1_-_92351613
|
1.391
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr3_+_32148000
|
1.370
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr3_+_238274
|
1.362
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr6_-_154831704
|
1.352
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr3_+_23987514
|
1.352
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr13_-_36705513
|
1.334
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr6_-_79787939
|
1.327
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr1_-_169455099
|
1.312
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr22_-_39240016
|
1.293
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr15_-_40401041
|
1.291
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr9_-_130742803
|
1.284
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr15_-_77712436
|
1.273
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr7_+_30174343
|
1.241
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr16_+_57126439
|
1.235
|
NM_152727
|
CPNE2
|
copine II
|
|
chr4_-_17812093
|
1.232
|
NM_017741
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr6_+_143999058
|
1.226
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr5_+_140345746
|
1.225
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr15_-_61521478
|
1.218
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_-_125933186
|
1.210
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr15_-_60884615
|
1.209
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_-_6494985
|
1.203
|
NM_001248007
NM_006458
NM_033278
|
TRIM3
|
tripartite motif containing 3
|
|
chr2_+_242641310
|
1.188
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr12_+_108523464
|
1.179
|
NM_014653
|
WSCD2
|
WSC domain containing 2
|
|
chr16_+_69599868
|
1.174
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr2_+_109065576
|
1.169
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr6_+_143929316
|
1.156
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr2_+_191273012
|
1.154
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr6_+_138483024
|
1.152
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr6_-_107435635
|
1.140
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr12_+_111843743
|
1.125
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr2_+_45878912
|
1.123
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr9_+_74729510
|
1.116
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chrX_+_16964730
|
1.108
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr11_+_111848008
|
1.099
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr5_+_140261792
|
1.096
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr1_+_164528586
|
1.075
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_+_118307072
|
1.074
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr20_+_30865435
|
1.070
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr1_-_57045235
|
1.069
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr7_-_152133071
|
1.067
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr9_-_99329092
|
1.056
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_-_92371558
|
1.055
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_179059071
|
1.053
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr3_-_53079285
|
1.041
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr16_-_3285069
|
1.040
|
NM_001145447
NM_003454
|
ZNF200
|
zinc finger protein 200
|
|
chr6_+_126070724
|
1.016
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr20_+_48429249
|
1.015
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr12_+_12764755
|
1.003
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr14_-_61190458
|
0.976
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr10_-_98346793
|
0.960
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr2_+_149402559
|
0.960
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr12_-_56652118
|
0.951
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr3_+_16926451
|
0.939
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr17_-_62502410
|
0.931
|
NM_004396
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr9_-_127703294
|
0.927
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr21_+_44394634
|
0.921
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr7_+_73442426
|
0.920
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr15_-_60919642
|
0.919
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_-_50316360
|
0.917
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr19_-_38085646
|
0.898
|
NM_016536
|
ZNF571
|
zinc finger protein 571
|
|
chr16_+_50775960
|
0.887
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr3_+_179370779
|
0.885
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr11_+_47290893
|
0.883
|
NM_130470
|
MADD
|
MAP-kinase activating death domain
|
|
chr2_-_45236521
|
0.874
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr2_-_202316227
|
0.868
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr3_-_36986535
|
0.866
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr11_+_8102691
|
0.866
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr3_-_53080038
|
0.864
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr6_+_43211221
|
0.864
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr14_-_39901619
|
0.862
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr9_+_91933384
|
0.861
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chrX_-_20284708
|
0.861
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr5_+_176237435
|
0.851
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr5_+_140180782
|
0.847
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr3_-_64211111
|
0.831
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr9_-_99382073
|
0.831
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr21_-_36421586
|
0.830
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_227505796
|
0.826
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr7_-_47621741
|
0.819
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr5_+_173315313
|
0.817
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr17_-_9940004
|
0.814
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr1_+_167189948
|
0.808
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr20_+_11898536
|
0.808
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr3_-_69101236
|
0.804
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr7_+_98476084
|
0.804
|
NM_001244580
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr9_-_130712866
|
0.802
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr1_+_26737232
|
0.801
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr17_-_9929567
|
0.796
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr16_+_50776012
|
0.795
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr15_-_31283617
|
0.790
|
NM_017762
|
MTMR10
|
myotubularin related protein 10
|
|
chr7_-_100286869
|
0.789
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr13_-_110438896
|
0.789
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr2_-_222436996
|
0.785
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr2_+_179184970
|
0.783
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr16_+_50186809
|
0.777
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr1_+_167298280
|
0.768
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr22_+_39853316
|
0.763
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr10_-_1779669
|
0.760
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr11_+_125034558
|
0.755
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr6_+_87865261
|
0.742
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr4_-_125633716
|
0.737
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr9_+_96338671
|
0.737
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr6_-_166075520
|
0.730
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr4_-_76555570
|
0.729
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr12_+_118814310
|
0.718
|
NM_022491
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae)
|
|
chr19_-_6110561
|
0.715
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr3_-_11761946
|
0.714
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr22_+_29168628
|
0.714
|
NM_173510
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr2_+_32582046
|
0.710
|
NM_016252
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr16_+_2525108
|
0.708
|
NM_001199107
NM_020705
|
TBC1D24
|
TBC1 domain family, member 24
|
|
chr9_+_74764266
|
0.707
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr10_+_69644938
|
0.703
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr11_+_111807857
|
0.695
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr10_+_129845806
|
0.692
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr5_+_140235594
|
0.690
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr15_+_41952572
|
0.689
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr3_-_118864881
|
0.687
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr3_-_11610264
|
0.686
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr17_-_10101854
|
0.681
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr2_-_97535669
|
0.680
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr9_-_128003634
|
0.679
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr15_-_51914770
|
0.677
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr9_+_129089092
|
0.674
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr14_+_90863302
|
0.666
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr18_-_72921280
|
0.666
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr4_+_48343502
|
0.658
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr8_+_86089618
|
0.656
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr2_-_164592512
|
0.655
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr1_+_26438267
|
0.654
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr7_-_6098813
|
0.653
|
NM_001134335
NM_014413
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1
|
|
chr3_+_16974581
|
0.649
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chrX_-_130037206
|
0.647
|
NM_006375
NM_182314
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2
|
|
chr17_+_61699786
|
0.646
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|