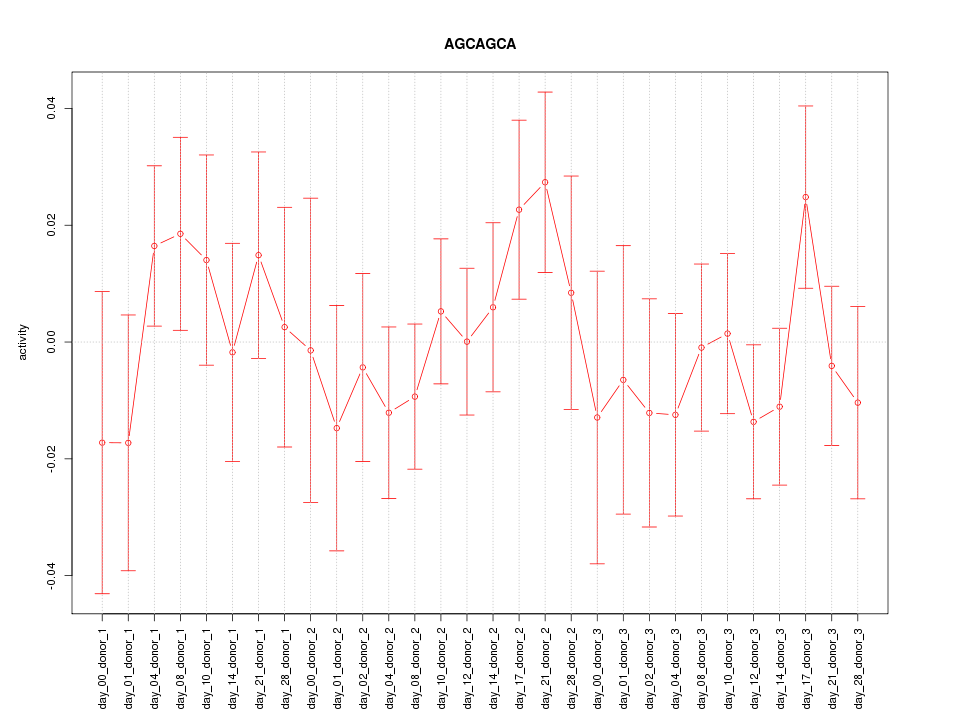
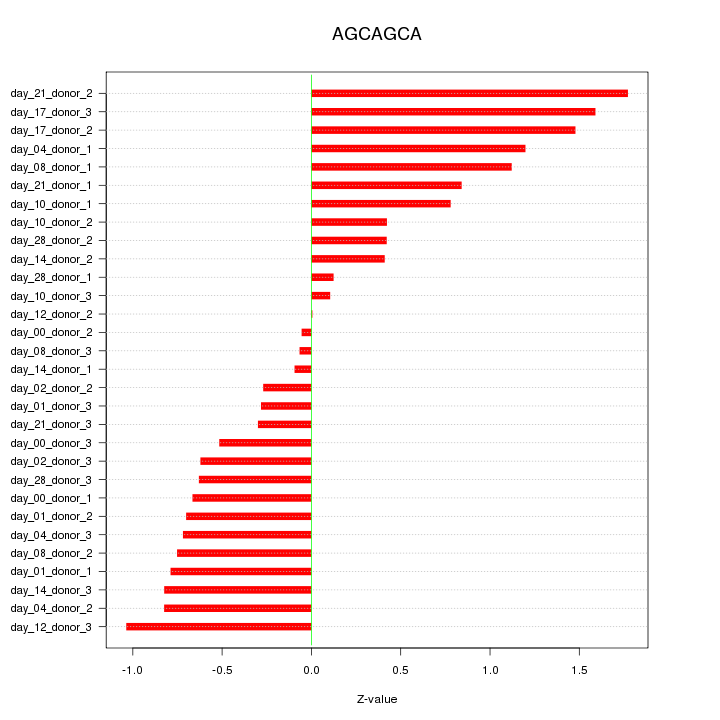
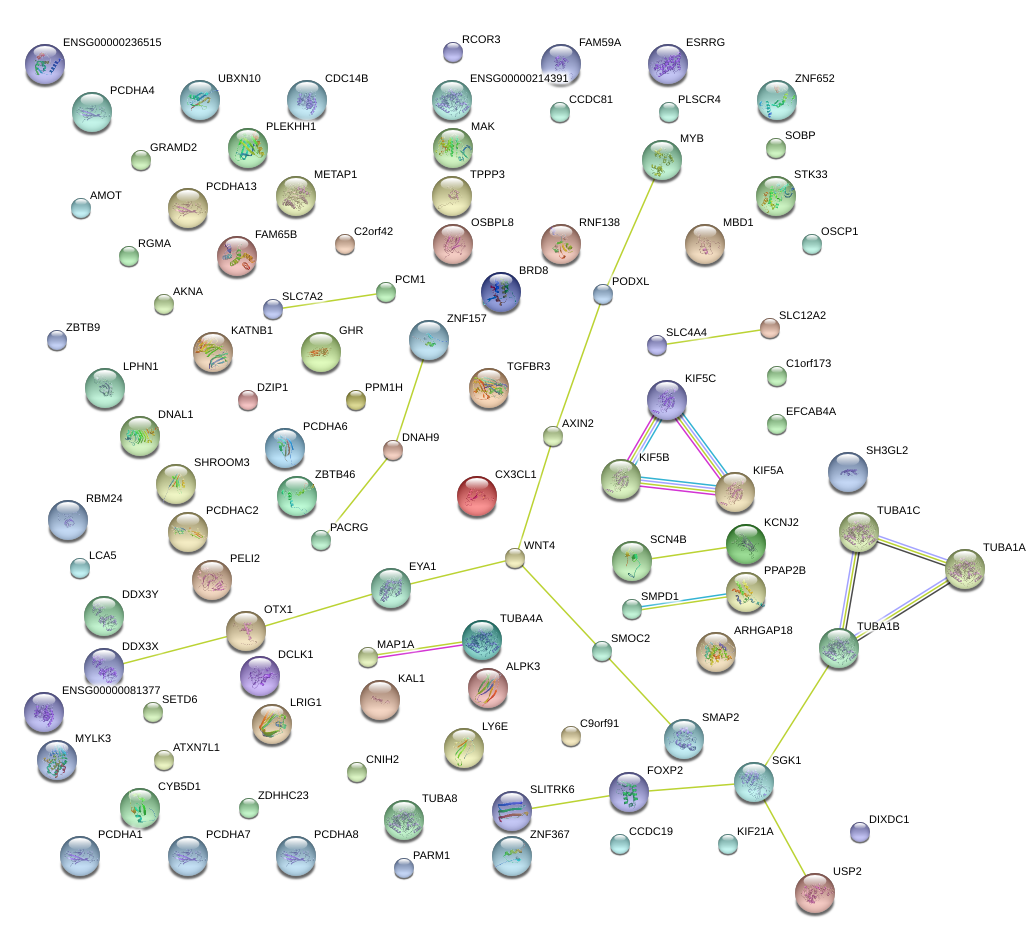
|
chr16_-_67427335
|
3.113
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr6_-_24911194
|
2.163
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr6_+_135502445
|
1.938
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_-_159869897
|
1.923
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr6_+_17281773
|
1.916
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
1.881
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282931
|
1.879
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr11_+_111848008
|
1.653
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr3_-_145968958
|
1.610
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr11_+_827584
|
1.472
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr12_-_49582800
|
1.420
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr8_+_17354562
|
1.419
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_+_20512577
|
1.378
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr16_+_57406398
|
1.349
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr4_+_72052354
|
1.298
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr15_+_43809805
|
1.275
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr5_+_140254930
|
1.239
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140213968
|
1.222
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr17_-_63557615
|
1.204
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr5_+_140186658
|
1.197
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_+_140220811
|
1.190
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr6_-_10838712
|
1.188
|
NM_001242385
NM_001242957
NM_005906
|
MAK
|
male germ cell-associated kinase
|
|
chr17_+_7761063
|
1.165
|
NM_144607
|
CYB5D1
|
cytochrome b5 domain containing 1
|
|
chr1_-_75139421
|
1.164
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr8_+_17396285
|
1.159
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr19_-_14316980
|
1.155
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr1_-_22469414
|
1.122
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr7_-_131241321
|
1.098
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr11_+_111807857
|
1.094
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr11_-_8615412
|
1.065
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr20_+_31823801
|
1.058
|
NM_001243193
NM_016583
NM_130852
|
BPIFA1
|
BPI fold containing family A, member 1
|
|
chr11_-_118016134
|
1.032
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr17_+_68165660
|
1.031
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr5_+_140247767
|
0.982
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr11_-_118023534
|
0.949
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chrY_+_15016018
|
0.924
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr4_+_77356252
|
0.921
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr14_+_56584854
|
0.912
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr3_-_66550844
|
0.911
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr4_+_75858205
|
0.903
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr11_-_119234859
|
0.891
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr12_-_63328663
|
0.883
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr20_-_62436855
|
0.880
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr14_+_67999915
|
0.878
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr12_-_39836760
|
0.877
|
NM_001173463
NM_001173464
NM_001173465
NM_017641
|
KIF21A
|
kinesin family member 21A
|
|
chr11_+_66045645
|
0.858
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr1_-_36915969
|
0.844
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr9_+_117373622
|
0.828
|
NM_153045
|
C9orf91
|
chromosome 9 open reading frame 91
|
|
chrX_-_112066353
|
0.810
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr5_+_140165875
|
0.791
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr1_-_217262946
|
0.782
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_-_80247103
|
0.779
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr4_+_72204769
|
0.772
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr13_-_36705513
|
0.760
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr7_+_114055048
|
0.759
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr5_+_140207562
|
0.753
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr16_-_46782104
|
0.745
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr8_+_144099900
|
0.744
|
NM_001127213
NM_002346
|
LY6E
|
lymphocyte antigen 6 complex, locus E
|
|
chr1_-_217311095
|
0.730
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_92351613
|
0.726
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_+_42423811
|
0.724
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr13_-_36429798
|
0.708
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr2_+_149632772
|
0.705
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr1_-_216896640
|
0.703
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_+_42424553
|
0.702
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr5_+_42565963
|
0.688
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr5_+_42548144
|
0.687
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr8_+_17780351
|
0.684
|
NM_006197
|
PCM1
|
pericentriolar material 1
|
|
chrX_-_8700170
|
0.679
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr11_+_86085777
|
0.679
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr5_+_42425093
|
0.679
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr5_+_42548314
|
0.677
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr16_+_57769626
|
0.677
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr2_-_70418086
|
0.674
|
NM_017880
|
C2orf42
|
chromosome 2 open reading frame 42
|
|
chr12_+_74931550
|
0.668
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr16_+_58549382
|
0.664
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr5_+_42565562
|
0.656
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr9_-_117156684
|
0.650
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr5_+_42550181
|
0.647
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr13_-_86373328
|
0.638
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr6_+_163149001
|
0.634
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr8_-_72268720
|
0.634
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr5_+_42549657
|
0.629
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr6_+_163148163
|
0.627
|
NM_001080378
NM_152410
|
PACRG
|
PARK2 co-regulated
|
|
chrY_+_15016698
|
0.626
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr18_+_29672568
|
0.608
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr11_-_119252364
|
0.605
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr6_-_134496006
|
0.601
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134497069
|
0.600
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_-_93632161
|
0.599
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr9_+_17578952
|
0.596
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr9_-_99329092
|
0.596
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr3_+_113666544
|
0.592
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr6_+_33422319
|
0.591
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr6_+_107811272
|
0.588
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_+_127419407
|
0.584
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr1_-_92371558
|
0.574
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr7_-_105319608
|
0.572
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr15_-_93616891
|
0.568
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr11_+_6411637
|
0.564
|
NM_000543
NM_001007593
|
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal
|
|
chr2_+_63277191
|
0.556
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr15_-_93617091
|
0.551
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr1_-_57045235
|
0.551
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr8_-_72274466
|
0.547
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr5_+_140345746
|
0.545
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr1_+_211433285
|
0.539
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr15_-_93616358
|
0.537
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_72490109
|
0.527
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr6_-_130031278
|
0.527
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr14_+_74111577
|
0.527
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chrX_-_112084006
|
0.526
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr13_-_96296919
|
0.524
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr6_-_134639195
|
0.519
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr18_-_30050394
|
0.518
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr17_-_47439326
|
0.517
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr1_+_40839368
|
0.515
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr5_+_67511578
|
0.508
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr15_-_93631751
|
0.508
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr6_-_134498973
|
0.506
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_27561006
|
0.506
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr2_+_63277936
|
0.505
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr8_+_79578258
|
0.502
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr20_-_1165116
|
0.501
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr1_+_40859016
|
0.500
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr5_+_140180782
|
0.500
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr5_+_67584251
|
0.491
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_-_89956531
|
0.489
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr8_+_81398416
|
0.487
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr19_-_7293876
|
0.482
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chrX_-_108976509
|
0.482
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr5_+_140261792
|
0.481
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr22_+_29279573
|
0.476
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr10_-_94050799
|
0.468
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr5_+_67586466
|
0.461
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_+_143999058
|
0.456
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr6_+_143929316
|
0.455
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_-_77459198
|
0.444
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr11_-_72852957
|
0.442
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr2_+_109065576
|
0.440
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr1_+_180601121
|
0.433
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr1_+_40840304
|
0.429
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr5_-_54830369
|
0.429
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr7_-_95225802
|
0.428
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr6_+_52285804
|
0.425
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr1_+_40862471
|
0.425
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr1_+_33722158
|
0.420
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr13_+_50589389
|
0.415
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr5_+_140235594
|
0.413
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr12_-_89918531
|
0.413
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr15_-_66084544
|
0.411
|
NM_001144823
NM_005848
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr12_+_122459791
|
0.411
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr2_-_122042767
|
0.410
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr12_+_12764755
|
0.407
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr5_+_67588395
|
0.405
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr18_+_29671817
|
0.400
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr14_-_39901619
|
0.399
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr1_+_179923870
|
0.395
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr13_-_30424795
|
0.395
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr11_-_61124223
|
0.394
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr10_-_94003002
|
0.388
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_99382073
|
0.388
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr6_+_80340971
|
0.384
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr6_-_170599568
|
0.381
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr16_+_10522724
|
0.379
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr9_-_126030854
|
0.377
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr18_+_67956136
|
0.375
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr4_+_184826427
|
0.374
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr1_+_211432693
|
0.374
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr7_-_152133071
|
0.373
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr9_+_4679557
|
0.373
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr10_+_88516340
|
0.370
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr11_+_100557851
|
0.369
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr16_+_67927170
|
0.369
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr12_+_7342269
|
0.366
|
NM_001131023
NM_001131024
NM_001131025
|
PEX5
|
peroxisomal biogenesis factor 5
|
|
chr1_+_109289266
|
0.366
|
NM_007269
|
STXBP3
|
syntaxin binding protein 3
|
|
chr7_+_75831194
|
0.365
|
NM_001110199
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr10_-_21463019
|
0.365
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr14_-_61190458
|
0.364
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr10_-_28034746
|
0.362
|
NM_001242702
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr19_-_46389307
|
0.359
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr20_+_30865435
|
0.355
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr6_-_79787939
|
0.352
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr11_+_6502607
|
0.350
|
NM_012192
|
FXC1
|
fracture callus 1 homolog (rat)
|
|
chr17_+_36861857
|
0.349
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr10_+_92980322
|
0.348
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr5_+_102201526
|
0.347
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr9_-_126030792
|
0.347
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr5_-_159739482
|
0.347
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr11_-_73693883
|
0.346
|
NM_003355
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr2_+_54952148
|
0.343
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr6_+_52284948
|
0.342
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr14_-_92506284
|
0.342
|
NM_004239
|
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr1_+_113615787
|
0.341
|
NM_014813
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr12_+_7342956
|
0.340
|
NM_000319
|
PEX5
|
peroxisomal biogenesis factor 5
|
|
chr20_-_40246991
|
0.338
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr3_-_107941168
|
0.335
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr5_+_167718485
|
0.335
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr11_-_110583120
|
0.332
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr20_+_278200
|
0.330
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr15_+_69452799
|
0.325
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|