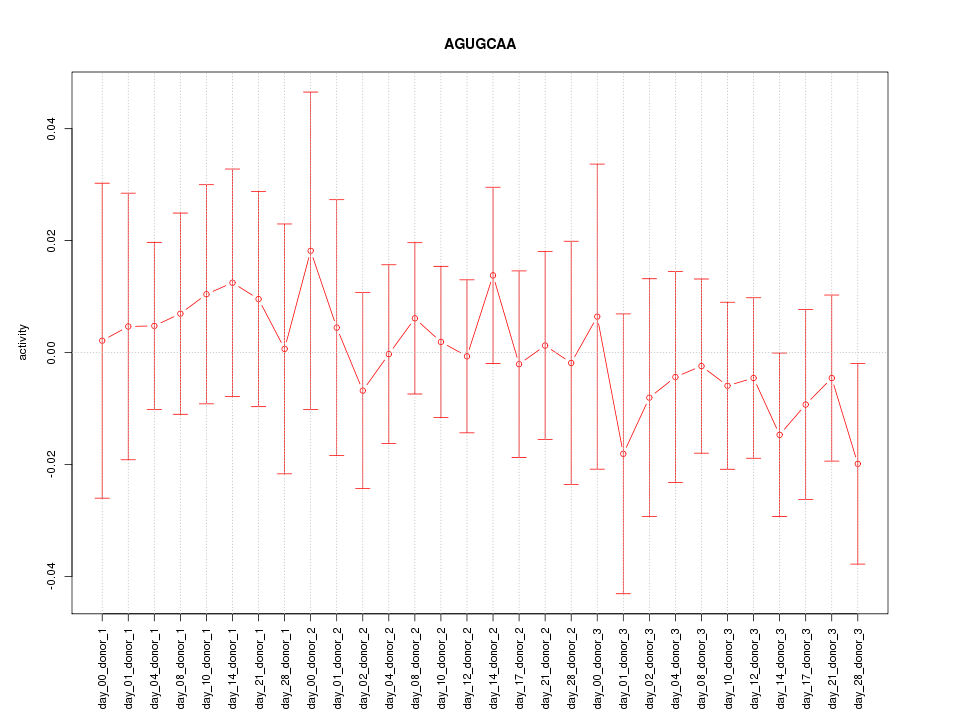
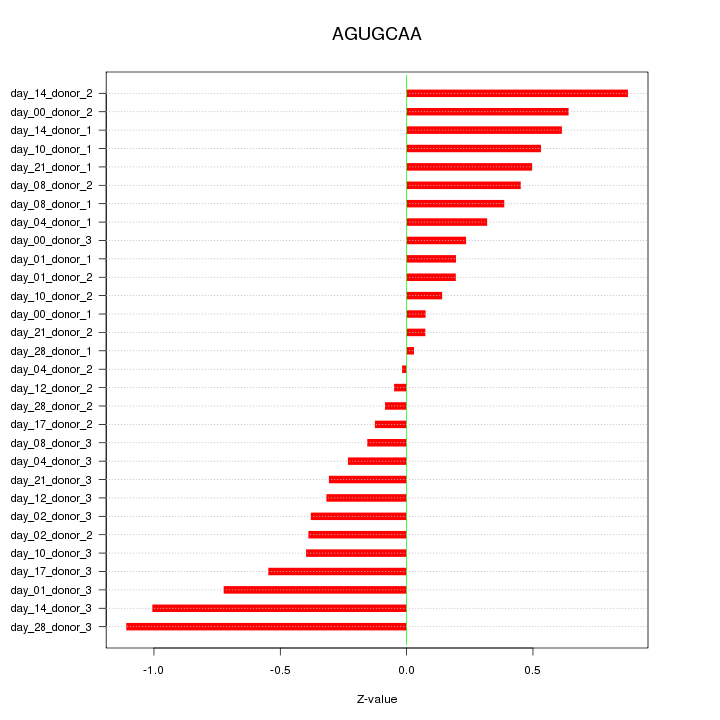
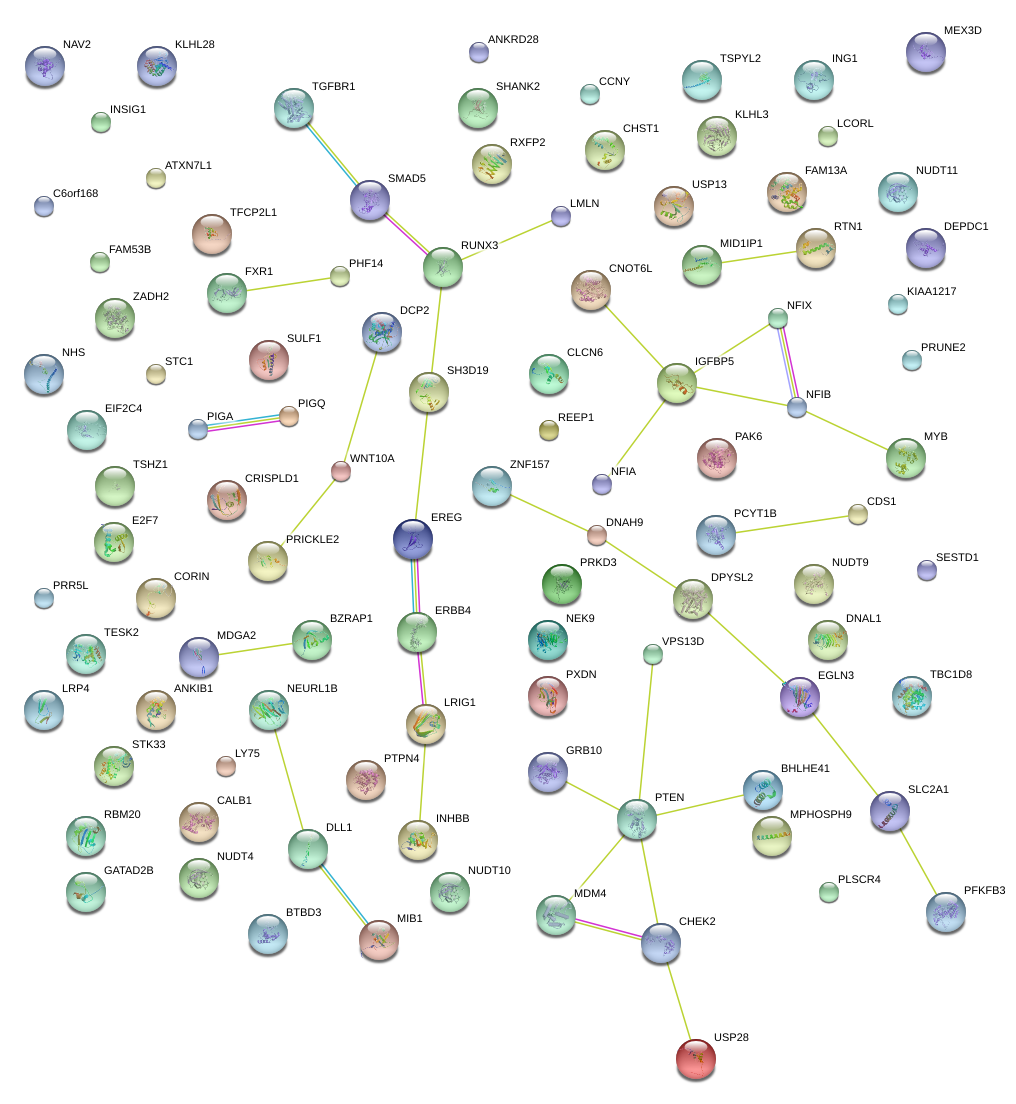
|
chr14_-_34419957
|
0.871
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr8_+_26371461
|
0.730
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_+_26435339
|
0.654
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_+_75896707
|
0.565
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr6_+_135502445
|
0.527
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr11_-_46939908
|
0.483
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr2_-_101767714
|
0.424
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr2_-_217560247
|
0.419
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr11_-_70507911
|
0.404
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr5_+_172068188
|
0.400
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr2_-_86565205
|
0.392
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr2_-_1748285
|
0.384
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr3_-_64211111
|
0.380
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr8_-_23712297
|
0.369
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_+_179370779
|
0.357
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr11_-_70935807
|
0.353
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_-_99797530
|
0.339
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr1_-_45956795
|
0.337
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr2_-_213403280
|
0.330
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr7_-_105319608
|
0.322
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chrX_+_17393537
|
0.322
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chrX_+_17653412
|
0.317
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr6_-_170599568
|
0.316
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr11_+_19734821
|
0.316
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr19_-_1567875
|
0.309
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr3_+_197687070
|
0.304
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr10_-_126432893
|
0.297
|
NM_014661
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr2_-_86564632
|
0.297
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr11_+_19372270
|
0.295
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr4_-_89744154
|
0.282
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr11_-_45687135
|
0.280
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr14_-_60097531
|
0.272
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr2_+_121103670
|
0.269
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr10_+_35535952
|
0.267
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr14_+_74111577
|
0.266
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr15_+_40532034
|
0.265
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr14_-_75593720
|
0.263
|
NM_033116
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr20_+_11898536
|
0.249
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr9_-_14398981
|
0.247
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_36422546
|
0.242
|
NM_001160169
|
PRR5L
|
proline rich 5 like
|
|
chr11_-_8615412
|
0.242
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr10_+_6244839
|
0.238
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr10_+_35625608
|
0.238
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr13_+_111367328
|
0.232
|
NM_005537
|
ING1
|
inhibitor of growth family, member 1
|
|
chr15_+_40509628
|
0.228
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr2_+_219745243
|
0.227
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr10_+_89623091
|
0.224
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr17_-_56406120
|
0.221
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr4_-_89978117
|
0.221
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr11_+_36397534
|
0.220
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr8_-_91094924
|
0.219
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr10_+_6186758
|
0.217
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr12_-_77459198
|
0.215
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr18_+_72922725
|
0.215
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr9_-_14314036
|
0.214
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr13_+_111365609
|
0.210
|
NM_198218
NM_198219
|
ING1
|
inhibitor of growth family, member 1
|
|
chr1_+_12290056
|
0.205
|
NM_015378
NM_018156
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr7_+_155089483
|
0.205
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr1_+_11866206
|
0.204
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr4_+_75230856
|
0.203
|
NM_001432
|
EREG
|
epiregulin
|
|
chr20_+_11871364
|
0.202
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr11_+_36317724
|
0.201
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr1_-_43424834
|
0.200
|
NM_006516
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr11_-_113746205
|
0.199
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr10_+_112404104
|
0.196
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr2_-_180129245
|
0.194
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr1_+_36273786
|
0.191
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr3_-_15900928
|
0.189
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr4_-_78740508
|
0.186
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr14_-_60337350
|
0.182
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr7_-_50772997
|
0.181
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr4_-_152147520
|
0.180
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chrX_-_15353675
|
0.176
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr12_-_26277807
|
0.175
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr1_-_25256767
|
0.174
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr4_+_85504056
|
0.174
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr7_+_11013453
|
0.172
|
NM_014660
|
PHF14
|
PHD finger protein 14
|
|
chr7_+_91875411
|
0.171
|
NM_019004
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1
|
|
chr1_-_68962726
|
0.170
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr3_-_15839674
|
0.169
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chrX_-_24665454
|
0.168
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr5_-_137071703
|
0.168
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr14_-_47812394
|
0.167
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr8_+_70378858
|
0.165
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr9_-_79520878
|
0.164
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr7_-_50861114
|
0.163
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_-_160761266
|
0.162
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr22_-_36424397
|
0.162
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_-_37544165
|
0.160
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr3_-_145968958
|
0.159
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chrX_+_51075082
|
0.159
|
NM_153183
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr12_-_123706393
|
0.158
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr5_+_135468522
|
0.158
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr9_+_101867401
|
0.157
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr18_-_72921280
|
0.157
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr2_+_120517206
|
0.156
|
NM_002830
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte)
|
|
chr14_-_45431121
|
0.156
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chrX_+_53111482
|
0.154
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr13_+_111364969
|
0.153
|
NM_198217
|
ING1
|
inhibitor of growth family, member 1
|
|
chr7_-_50800018
|
0.152
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr3_+_180630054
|
0.152
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr2_-_122042767
|
0.150
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr3_-_66550844
|
0.150
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr4_-_18023358
|
0.148
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr1_+_204485461
|
0.148
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr5_+_112312392
|
0.147
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chrX_+_38663072
|
0.146
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr18_+_19321544
|
0.146
|
NM_020774
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr1_-_153895450
|
0.144
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr13_+_32313678
|
0.140
|
NM_001166058
NM_130806
|
RXFP2
|
relaxin/insulin-like family peptide receptor 2
|
|
chr10_+_24497719
|
0.139
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr12_-_120315074
|
0.138
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr6_+_69344939
|
0.138
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr12_+_109915430
|
0.137
|
NM_130466
NM_183415
|
UBE3B
|
ubiquitin protein ligase E3B
|
|
chr8_-_92053043
|
0.136
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr2_+_179184970
|
0.136
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_+_244214552
|
0.136
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr12_+_56414688
|
0.136
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr7_+_114562140
|
0.135
|
NM_001166345
NM_001166346
NM_199072
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr15_-_64648359
|
0.135
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr1_+_244216506
|
0.134
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr7_-_106301329
|
0.133
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr4_+_129730772
|
0.131
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr3_+_16926451
|
0.130
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr6_+_64356430
|
0.130
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr10_-_94050799
|
0.130
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_-_15798057
|
0.129
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr5_-_58295758
|
0.128
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_7396030
|
0.127
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr2_+_131674223
|
0.126
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr22_+_38004914
|
0.126
|
NM_001172688
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr15_-_49338496
|
0.125
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr18_+_13620866
|
0.125
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr1_-_169336999
|
0.124
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr22_+_38004454
|
0.123
|
NM_001001560
NM_001001561
NM_001172687
NM_013365
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr12_+_120105656
|
0.123
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr3_+_140950671
|
0.122
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr3_+_61547179
|
0.122
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr3_+_12392993
|
0.122
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_+_97889467
|
0.120
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr18_+_13218777
|
0.118
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr12_+_104458235
|
0.118
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr3_+_4535030
|
0.118
|
NM_001099952
NM_001168272
NM_002222
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr5_-_59189620
|
0.118
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_52978022
|
0.117
|
NM_005486
|
TOM1L1
|
target of myb1 (chicken)-like 1
|
|
chr18_+_13611664
|
0.116
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr6_+_46097700
|
0.116
|
NM_014936
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr2_+_203241044
|
0.115
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr7_+_23636970
|
0.114
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr1_+_26438267
|
0.114
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr16_-_23521803
|
0.113
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr10_+_23983674
|
0.113
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr4_-_141075232
|
0.113
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr2_+_173420776
|
0.112
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr18_-_53255734
|
0.111
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr11_+_120207617
|
0.111
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr12_-_50419160
|
0.110
|
NM_001126103
NM_001126104
NM_013277
|
RACGAP1
|
Rac GTPase activating protein 1
|
|
chr5_-_58571916
|
0.110
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_89825292
|
0.108
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr1_-_243418675
|
0.108
|
NM_001042404
NM_001042405
NM_014812
|
CEP170
|
centrosomal protein 170kDa
|
|
chr11_-_10315680
|
0.108
|
NM_030962
|
SBF2
|
SET binding factor 2
|
|
chr6_-_138893666
|
0.106
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr5_+_56471272
|
0.105
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr19_+_10828725
|
0.105
|
NM_001005360
NM_001005361
NM_001005362
NM_001190716
NM_004945
|
DNM2
|
dynamin 2
|
|
chr1_+_185014449
|
0.104
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chr3_+_12329332
|
0.104
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_-_104178900
|
0.104
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr20_-_52199635
|
0.103
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr5_-_59783885
|
0.102
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_+_70405027
|
0.102
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr6_+_131571298
|
0.101
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr8_+_27491460
|
0.101
|
NM_016240
NM_182826
|
SCARA3
|
scavenger receptor class A, member 3
|
|
chr5_+_149151502
|
0.101
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr12_+_1100352
|
0.101
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr7_+_29603388
|
0.100
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr20_-_17949067
|
0.100
|
NM_014426
|
SNX5
|
sorting nexin 5
|
|
chr4_+_15004297
|
0.100
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr2_-_55646946
|
0.099
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr20_+_34742656
|
0.098
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr11_-_102826433
|
0.098
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr3_-_52931595
|
0.097
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr3_+_16974581
|
0.097
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chr4_+_106816593
|
0.096
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr1_-_212004113
|
0.096
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr3_+_14444083
|
0.096
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chrX_-_108976509
|
0.096
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chrX_+_38660653
|
0.095
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr7_+_115850546
|
0.095
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr6_-_42419782
|
0.094
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr15_-_23086842
|
0.092
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr10_+_31610063
|
0.092
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_96162185
|
0.092
|
NM_015188
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr3_+_120627026
|
0.092
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chrX_-_24690740
|
0.091
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr5_+_86564727
|
0.091
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr13_-_95364248
|
0.090
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr5_-_58334936
|
0.090
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_177053306
|
0.089
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr10_-_43762366
|
0.089
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr5_+_149109814
|
0.089
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|