|
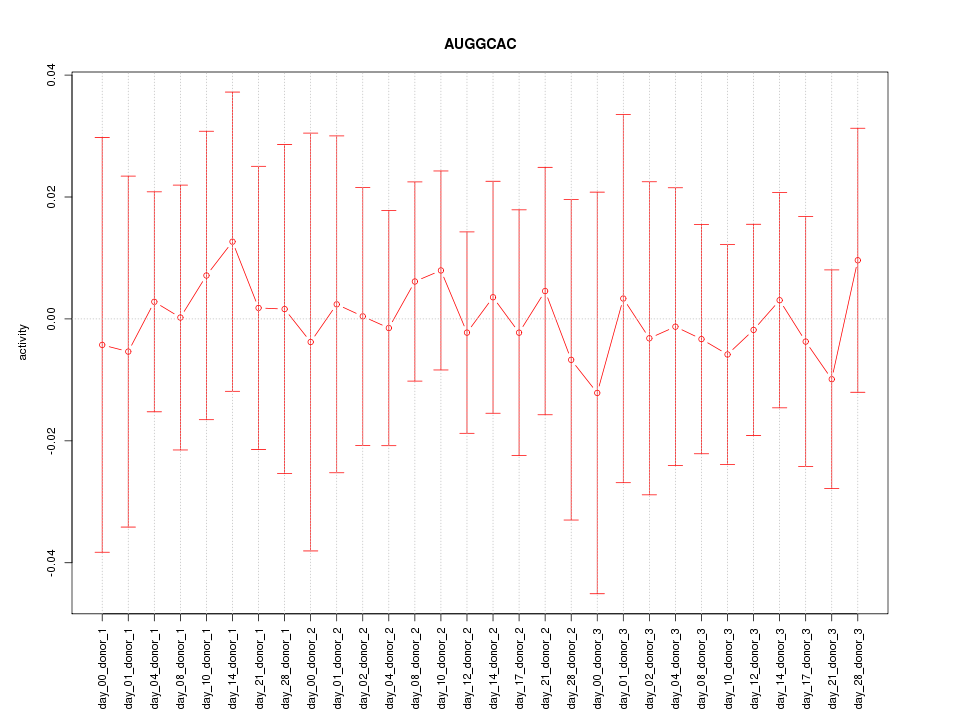
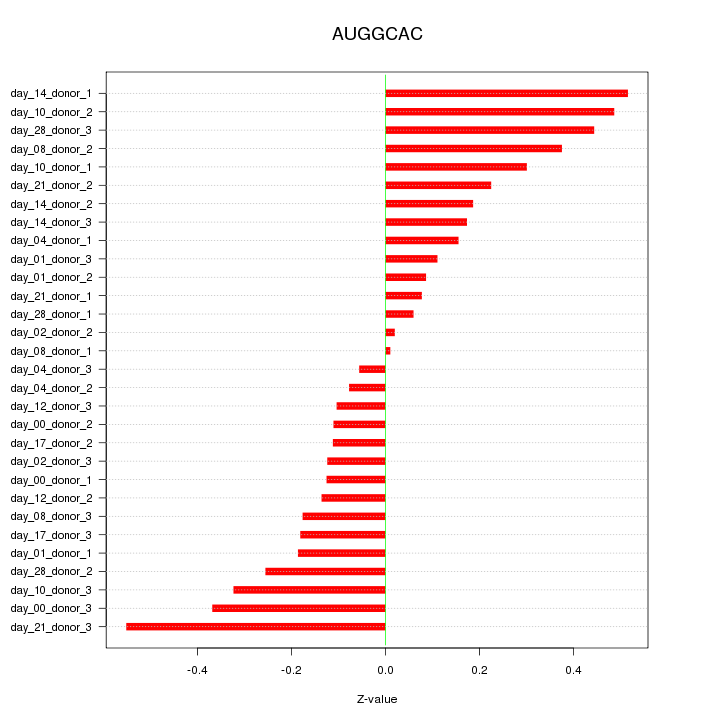
chr16_+_57406398
|
0.579
|
NM_002996
|
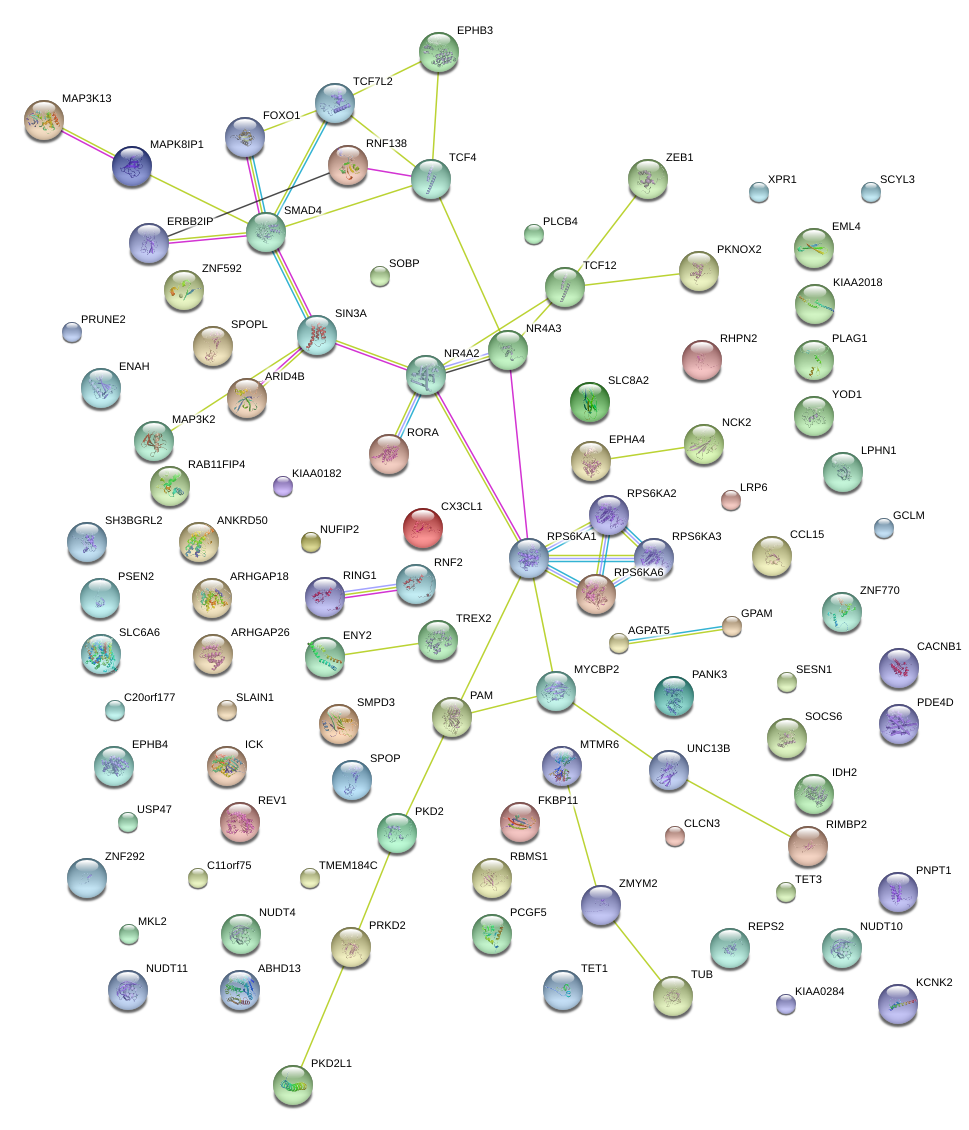
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr11_-_93276513
|
0.361
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chr11_+_45907046
|
0.345
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr6_-_130031278
|
0.215
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chrX_+_16964730
|
0.206
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr19_-_14316980
|
0.193
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr19_-_33555757
|
0.185
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr12_+_93771607
|
0.174
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr3_+_14444083
|
0.172
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr1_-_94374986
|
0.159
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr18_-_53255734
|
0.156
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr17_+_29718641
|
0.152
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr1_+_180601121
|
0.140
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr1_+_227058241
|
0.139
|
NM_000447
NM_012486
|
PSEN2
|
presenilin 2 (Alzheimer disease 4)
|
|
chr10_-_113943467
|
0.138
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr2_-_157189040
|
0.137
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chrX_-_20284708
|
0.134
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr10_+_70320044
|
0.134
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr18_+_67956136
|
0.128
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr9_-_79520878
|
0.124
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_-_169863048
|
0.124
|
NM_020423
NM_181093
|
SCYL3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr2_-_55920947
|
0.112
|
NM_033109
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1
|
|
chr2_-_161350304
|
0.107
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr16_-_68482373
|
0.104
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr15_-_75743776
|
0.104
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr18_+_29672568
|
0.103
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr15_-_75744029
|
0.102
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr15_-_75748123
|
0.099
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr15_-_60884615
|
0.099
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_-_12419702
|
0.098
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr5_+_65222381
|
0.097
|
NM_001006600
NM_001253697
NM_001253698
NM_001253699
NM_001253701
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr15_-_61521478
|
0.096
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_+_110346549
|
0.095
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr13_-_25861608
|
0.093
|
NM_004685
|
MTMR6
|
myotubularin related protein 6
|
|
chr5_+_142150291
|
0.093
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr20_+_9049660
|
0.091
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr12_-_131002409
|
0.091
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr18_+_48556483
|
0.091
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr10_+_31610063
|
0.090
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr16_+_85645014
|
0.088
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr20_+_58515408
|
0.088
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr14_+_105331581
|
0.087
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr13_+_108870713
|
0.086
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr20_+_9288446
|
0.086
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr2_+_106361404
|
0.084
|
NM_001004722
NM_003581
|
NCK2
|
NCK adaptor protein 2
|
|
chr15_-_60919642
|
0.084
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_57210823
|
0.083
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr8_+_6565870
|
0.083
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr13_-_41240607
|
0.079
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr4_+_148538508
|
0.079
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr4_+_88928797
|
0.078
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr20_+_9198036
|
0.078
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr6_-_52926588
|
0.076
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr13_+_78271820
|
0.075
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr6_+_80340971
|
0.075
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr4_-_125633716
|
0.074
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr2_-_100106478
|
0.074
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr2_-_222436996
|
0.073
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr1_-_225840660
|
0.071
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr15_-_90645704
|
0.070
|
NM_002168
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chr13_+_20532906
|
0.070
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr1_+_215178884
|
0.069
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr11_+_11862955
|
0.069
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr11_+_8102691
|
0.068
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr2_+_106468203
|
0.068
|
NM_001004720
|
NCK2
|
NCK adaptor protein 2
|
|
chr1_-_207224298
|
0.068
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr3_-_113415313
|
0.068
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr19_-_47975244
|
0.064
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr1_+_215256559
|
0.064
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr6_+_107811272
|
0.064
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr2_+_139259349
|
0.064
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr20_+_58508818
|
0.063
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr3_+_185000714
|
0.063
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr1_+_185014449
|
0.061
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chr5_-_59783885
|
0.059
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_+_92980322
|
0.059
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr2_+_42396392
|
0.059
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr4_+_170541656
|
0.059
|
NM_001243372
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr6_-_109415514
|
0.058
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr16_+_85646923
|
0.058
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr2_+_74273449
|
0.058
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr13_+_20532788
|
0.057
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr17_-_27621163
|
0.056
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr5_+_102201526
|
0.052
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_-_168006413
|
0.052
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr6_+_87865261
|
0.052
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr8_-_57123858
|
0.051
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr6_-_109330190
|
0.051
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr15_-_35280496
|
0.051
|
NM_014106
|
ZNF770
|
zinc finger protein 770
|
|
chr15_+_85291802
|
0.050
|
NM_014630
|
ZNF592
|
zinc finger protein 592
|
|
chr9_+_35161950
|
0.050
|
NM_006377
|
UNC13B
|
unc-13 homolog B (C. elegans)
|
|
chr3_+_185080835
|
0.050
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr2_-_128100673
|
0.049
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr11_+_125034558
|
0.049
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr15_+_57511616
|
0.047
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_-_235490801
|
0.046
|
NM_001206794
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr16_+_14164879
|
0.046
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr18_-_47721165
|
0.046
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr14_-_68283293
|
0.045
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr10_+_22610138
|
0.045
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr1_-_70820363
|
0.045
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr2_+_219263060
|
0.045
|
NM_001206878
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr2_+_159313466
|
0.044
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr10_+_31608097
|
0.043
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr14_+_36295515
|
0.043
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr3_+_150804675
|
0.042
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr6_-_109330752
|
0.042
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr2_-_60780404
|
0.042
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr2_+_219264477
|
0.042
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr9_-_98270321
|
0.041
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr4_+_40058507
|
0.041
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr14_+_90863302
|
0.040
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr9_-_80646150
|
0.040
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr12_+_79258448
|
0.039
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr6_-_159239269
|
0.039
|
NM_003379
|
EZR
|
ezrin
|
|
chr3_+_140660639
|
0.039
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr14_+_102228134
|
0.039
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr6_-_62995852
|
0.038
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr22_-_21356351
|
0.038
|
NM_001008695
NM_030573
|
THAP7
|
THAP domain containing 7
|
|
chr5_-_58295758
|
0.037
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_70671331
|
0.036
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr9_-_98279246
|
0.036
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr7_+_20370724
|
0.036
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr17_-_37607314
|
0.035
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr12_+_64798050
|
0.035
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr4_+_15004297
|
0.034
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr8_-_24814028
|
0.033
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr8_+_120220554
|
0.033
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr18_+_29671817
|
0.033
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr5_-_58571916
|
0.033
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_69063768
|
0.032
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr11_+_63655986
|
0.031
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr7_-_16685360
|
0.031
|
NM_020319
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2
|
|
chr17_-_16118789
|
0.031
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr9_+_132934685
|
0.031
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr1_-_93257950
|
0.030
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr13_+_78315294
|
0.030
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr5_-_59189620
|
0.030
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58334936
|
0.029
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_53342320
|
0.029
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr7_-_8301767
|
0.029
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr10_+_114709963
|
0.029
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr17_-_60142598
|
0.028
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr9_-_98269686
|
0.028
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr12_+_79257772
|
0.028
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr9_-_98270830
|
0.028
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr7_+_101460881
|
0.028
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr21_-_27945267
|
0.027
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr5_-_32174397
|
0.027
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr9_-_98269480
|
0.027
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr17_+_27717942
|
0.027
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr12_+_27397077
|
0.026
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr11_-_60928878
|
0.026
|
NM_017966
|
VPS37C
|
vacuolar protein sorting 37 homolog C (S. cerevisiae)
|
|
chr7_+_35840595
|
0.023
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr1_-_153895450
|
0.023
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr8_-_141645603
|
0.023
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr11_-_116968986
|
0.023
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr3_-_71632741
|
0.022
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr1_-_235491494
|
0.021
|
NM_016374
NM_031371
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr9_-_135819995
|
0.021
|
NM_000368
NM_001162426
NM_001162427
|
TSC1
|
tuberous sclerosis 1
|
|
chr4_-_2263706
|
0.021
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr2_+_46769821
|
0.020
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr10_-_114206640
|
0.020
|
NM_022494
|
ZDHHC6
|
zinc finger, DHHC-type containing 6
|
|
chr3_+_38495778
|
0.020
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr4_-_129209983
|
0.019
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr9_-_114247024
|
0.019
|
NM_001080398
|
KIAA0368
|
KIAA0368
|
|
chr12_+_79439432
|
0.018
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr20_+_3451615
|
0.018
|
NM_001207047
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr1_-_67896098
|
0.018
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr11_+_12695851
|
0.017
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr1_-_151798552
|
0.017
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr12_-_16760935
|
0.017
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_8302141
|
0.016
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr5_+_153418518
|
0.015
|
NM_001135037
NM_001242336
NM_005927
|
MFAP3
|
microfibrillar-associated protein 3
|
|
chr10_-_25012543
|
0.015
|
NM_020824
|
ARHGAP21
|
Rho GTPase activating protein 21
|
|
chr7_-_71802207
|
0.014
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr11_-_47198675
|
0.014
|
NM_001242832
NM_032389
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr1_-_160254923
|
0.014
|
NM_001193644
NM_002857
|
PEX19
|
peroxisomal biogenesis factor 19
|
|
chr5_-_137071703
|
0.014
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr2_+_206547131
|
0.013
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr12_-_16759531
|
0.013
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_16097882
|
0.013
|
NM_001190438
NM_001190440
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr7_-_37956515
|
0.013
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr1_-_236767787
|
0.012
|
NM_018072
|
HEATR1
|
HEAT repeat containing 1
|
|
chr18_+_32073245
|
0.012
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr19_+_5623045
|
0.011
|
NM_001201338
NM_001201339
NM_001201340
NM_002967
|
SAFB
|
scaffold attachment factor B
|
|
chr6_-_3445198
|
0.011
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr1_+_204485461
|
0.011
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr6_+_88032289
|
0.011
|
NM_001042493
NM_020425
|
C6orf162
|
chromosome 6 open reading frame 162
|
|
chr4_-_11430502
|
0.010
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr4_-_78740508
|
0.010
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr17_-_74733419
|
0.010
|
NM_001195427
NM_003016
|
SRSF2
|
serine/arginine-rich splicing factor 2
|
|
chr5_+_271700
|
0.010
|
NM_013232
|
PDCD6
|
programmed cell death 6
|
|
chr12_+_72148653
|
0.010
|
NM_014999
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr10_-_75910525
|
0.009
|
NM_012095
NM_207012
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr16_+_9185534
|
0.009
|
NM_014117
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr6_-_132834165
|
0.009
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr10_+_22605300
|
0.007
|
NM_001204062
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough
COMM domain containing 3
|
|
chr9_+_129622935
|
0.007
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr6_+_131148544
|
0.007
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|