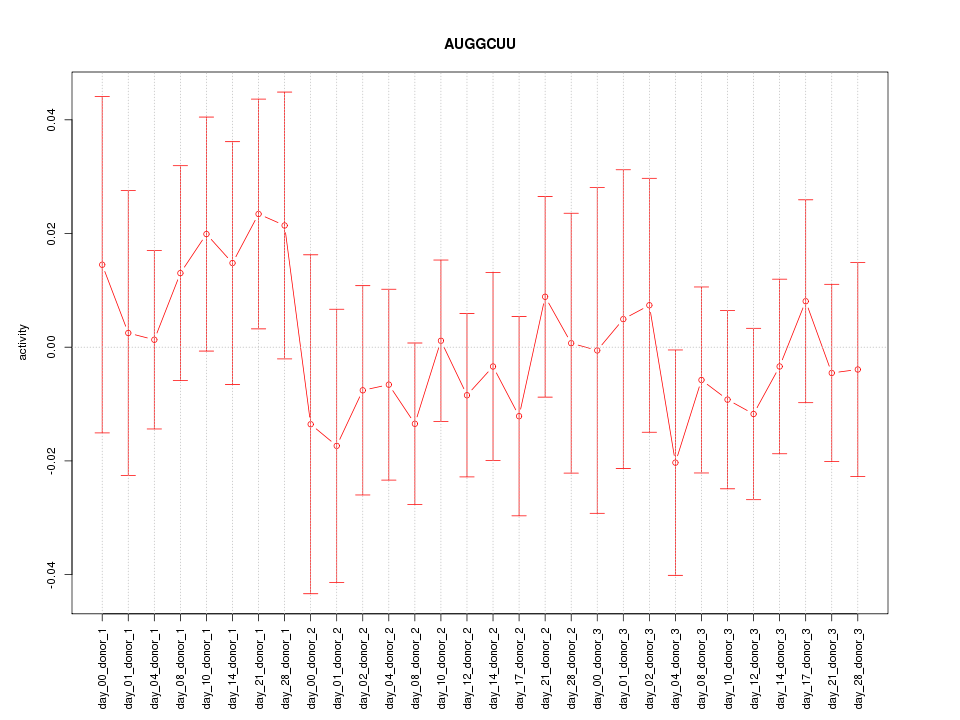
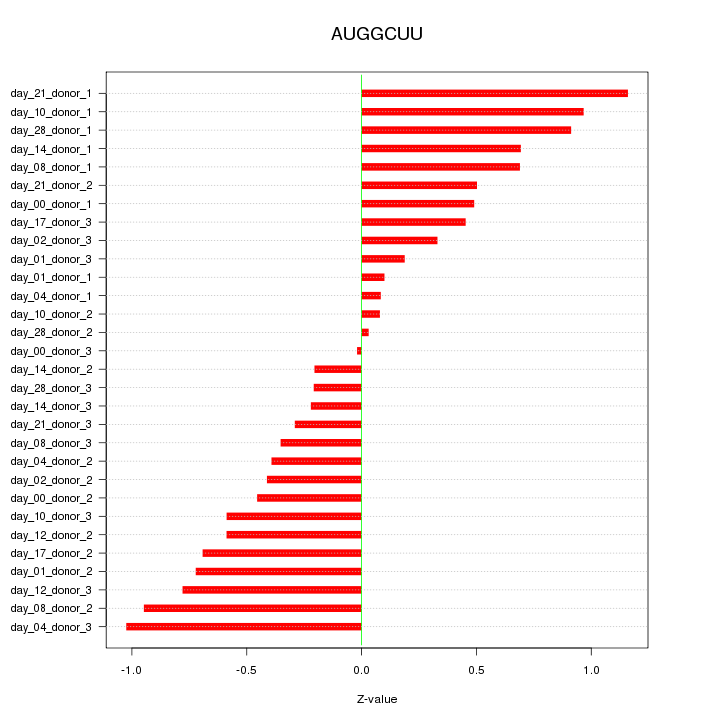
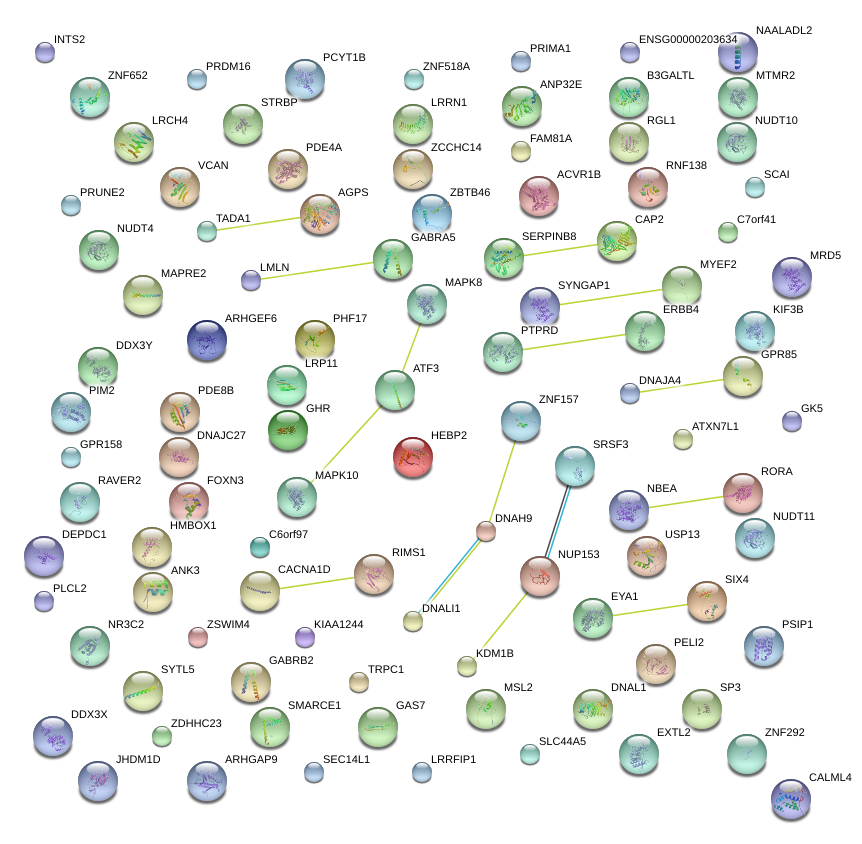
|
chr15_-_68498373
|
1.095
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr2_-_213403280
|
0.821
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr4_-_149363498
|
0.811
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr6_+_151815174
|
0.810
|
NM_025059
|
C6orf97
|
chromosome 6 open reading frame 97
|
|
chr13_+_36050885
|
0.713
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr1_+_183605207
|
0.701
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_+_18155535
|
0.683
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr15_-_48470453
|
0.640
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr13_+_35516390
|
0.598
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chrX_-_135863502
|
0.558
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_24665454
|
0.552
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr6_+_17393610
|
0.535
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr15_-_60884615
|
0.534
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_+_212738675
|
0.520
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr3_+_113666544
|
0.517
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr18_+_29672568
|
0.497
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr1_-_76076762
|
0.496
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr7_+_30174343
|
0.496
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr13_+_31774087
|
0.479
|
NM_194318
|
B3GALTL
|
beta 1,3-galactosyltransferase-like
|
|
chr3_+_197687070
|
0.461
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr6_-_150185370
|
0.454
|
NM_032832
|
LRP11
|
low density lipoprotein receptor-related protein 11
|
|
chrX_+_37865834
|
0.452
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr1_+_212781969
|
0.452
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr19_+_13906207
|
0.448
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chrX_+_37892786
|
0.443
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chrY_+_15016018
|
0.435
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr17_-_10101854
|
0.435
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_9940004
|
0.430
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_9929567
|
0.416
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr3_+_179370779
|
0.415
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr6_-_119031230
|
0.408
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chrX_-_48776282
|
0.408
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr15_-_61521478
|
0.396
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr14_+_74111577
|
0.388
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr3_+_3841079
|
0.382
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr14_+_56584854
|
0.376
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr6_+_87865261
|
0.375
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr9_-_79520878
|
0.375
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr17_-_47439326
|
0.372
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr2_+_238536183
|
0.371
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr8_-_72268720
|
0.365
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_-_95657255
|
0.353
|
NM_001243571
NM_016156
NM_201278
NM_201281
|
MTMR2
|
myotubularin related protein 2
|
|
chr6_+_72926462
|
0.347
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_-_60919642
|
0.343
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_+_10543110
|
0.342
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr3_-_135914654
|
0.340
|
NM_018133
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr18_+_29671817
|
0.333
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr5_+_76506705
|
0.322
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr6_+_138483024
|
0.321
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr9_-_15510899
|
0.319
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr8_-_72274466
|
0.319
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr18_+_32556891
|
0.317
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_-_166845533
|
0.313
|
NM_053053
|
TADA1
|
transcriptional adaptor 1
|
|
chr10_-_62149487
|
0.310
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_38022514
|
0.310
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr10_-_61900659
|
0.308
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrY_+_15016698
|
0.307
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr17_-_60005309
|
0.307
|
NM_020748
|
INTS2
|
integrator complex subunit 2
|
|
chr9_-_126030854
|
0.306
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr14_-_94254765
|
0.306
|
NM_178013
|
PRIMA1
|
proline rich membrane anchor 1
|
|
chr8_+_28747885
|
0.304
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr20_+_30865435
|
0.300
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr10_-_62493282
|
0.299
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_105319608
|
0.297
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr1_-_101360405
|
0.297
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr9_-_126030792
|
0.296
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr19_+_10563581
|
0.292
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr10_-_62332666
|
0.290
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr15_+_78556901
|
0.288
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr3_+_142443214
|
0.288
|
NM_001251845
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr18_+_32558207
|
0.288
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr15_+_78558522
|
0.285
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_+_59730272
|
0.280
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr12_+_93771607
|
0.277
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr15_+_78556485
|
0.270
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr19_+_10531128
|
0.269
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr6_+_36562089
|
0.269
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr7_-_139876718
|
0.268
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr10_+_97889467
|
0.265
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr4_+_129730772
|
0.261
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr3_+_16926451
|
0.255
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr1_+_212788359
|
0.254
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr19_+_10541334
|
0.252
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr3_+_53529030
|
0.247
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr8_+_28748030
|
0.246
|
NM_024567
|
HMBOX1
|
homeobox containing 1
|
|
chr1_+_65210724
|
0.239
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr20_-_62436855
|
0.238
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr15_+_27112106
|
0.237
|
NM_001165037
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr6_+_33387846
|
0.235
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr7_-_112726143
|
0.235
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_+_82767492
|
0.233
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr3_-_141944441
|
0.230
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr1_+_2985730
|
0.229
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr3_+_174577069
|
0.227
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr17_+_75085234
|
0.225
|
NM_001204410
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr2_-_174828775
|
0.223
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr6_+_72922513
|
0.222
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_+_27111865
|
0.220
|
NM_000810
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chrX_-_24690740
|
0.216
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr14_-_90085325
|
0.216
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr16_+_22019438
|
0.210
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr3_-_135913276
|
0.208
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr14_-_61190458
|
0.207
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr9_-_10612722
|
0.207
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr2_-_25194772
|
0.207
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr17_-_38804037
|
0.203
|
NM_003079
|
SMARCE1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1
|
|
chr6_-_17706617
|
0.203
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr9_-_8733893
|
0.202
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr17_-_9862765
|
0.201
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr2_+_178257371
|
0.201
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr10_+_25464289
|
0.200
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr5_+_42423811
|
0.200
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr16_-_87525317
|
0.198
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr6_+_72596648
|
0.197
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_68962726
|
0.196
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr5_-_160975124
|
0.196
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr6_+_72926144
|
0.193
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_150208462
|
0.193
|
NM_001136478
NM_030920
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr3_+_16974581
|
0.193
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chr12_+_52345437
|
0.192
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr4_-_87374282
|
0.190
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr9_-_127905715
|
0.189
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr7_+_152456820
|
0.187
|
NM_001040135
NM_020445
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast)
|
|
chr6_-_116381765
|
0.186
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr6_+_144612872
|
0.185
|
NM_007124
|
UTRN
|
utrophin
|
|
chr14_-_95786095
|
0.185
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr10_+_101419169
|
0.184
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr10_-_75173807
|
0.184
|
NM_001156
NM_004034
|
ANXA7
|
annexin A7
|
|
chr18_-_29523087
|
0.184
|
NM_014939
|
TRAPPC8
|
trafficking protein particle complex 8
|
|
chr4_-_47916498
|
0.183
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr6_-_118973019
|
0.183
|
NM_001042475
NM_206921
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr15_-_40857229
|
0.183
|
NM_001080791
NM_001080792
NM_052849
|
C15orf57
|
chromosome 15 open reading frame 57
|
|
chr17_+_26646120
|
0.182
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr19_+_2328628
|
0.181
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr8_+_58906968
|
0.178
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr5_+_42565963
|
0.176
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr11_-_101454658
|
0.176
|
NM_004621
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6
|
|
chr5_+_42548144
|
0.176
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr10_+_69644938
|
0.175
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr5_+_42424553
|
0.175
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr12_+_111471827
|
0.173
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr14_-_45431121
|
0.172
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr5_+_112312392
|
0.171
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr11_-_115375065
|
0.171
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr2_-_240322620
|
0.171
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr11_-_130184459
|
0.169
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr5_+_42548314
|
0.169
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr5_+_42425093
|
0.168
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr4_-_87028805
|
0.168
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr17_+_75181294
|
0.167
|
NM_001144001
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr18_+_48556483
|
0.167
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr22_+_29279573
|
0.165
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr6_+_143999058
|
0.165
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_+_65218351
|
0.162
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr11_+_121322848
|
0.160
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr18_+_42260774
|
0.158
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr1_+_179923870
|
0.158
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr22_-_41842853
|
0.157
|
NM_016272
|
TOB2
|
transducer of ERBB2, 2
|
|
chr2_+_189156395
|
0.156
|
NM_001252668
NM_001252669
NM_016315
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr15_+_42066631
|
0.156
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr16_-_88772732
|
0.155
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr1_-_111682767
|
0.155
|
NM_178454
|
DRAM2
|
DNA-damage regulated autophagy modulator 2
|
|
chr12_+_19592311
|
0.153
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr4_+_154125589
|
0.153
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr10_+_114709963
|
0.152
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr15_+_77713229
|
0.151
|
NM_018200
|
HMG20A
|
high mobility group 20A
|
|
chr17_+_75136985
|
0.151
|
NM_001039573
NM_001143998
NM_001143999
NM_003003
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr2_+_191513832
|
0.151
|
NM_005966
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1)
|
|
chr16_-_88772807
|
0.150
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr5_+_42565562
|
0.150
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr20_+_43160435
|
0.149
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr10_+_69644341
|
0.149
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr3_+_97483563
|
0.148
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr17_+_27071008
|
0.148
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr1_-_93257950
|
0.147
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr21_-_43373938
|
0.146
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr3_-_27763784
|
0.146
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr11_-_18343670
|
0.145
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr17_-_19770988
|
0.145
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr4_+_154074269
|
0.145
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_-_77734285
|
0.144
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr6_+_137143683
|
0.143
|
NM_000288
|
PEX7
|
peroxisomal biogenesis factor 7
|
|
chr7_-_112727832
|
0.143
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr10_+_76586299
|
0.142
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr4_-_18023358
|
0.142
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr1_-_150207025
|
0.142
|
NM_001136479
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr4_+_71768056
|
0.140
|
NM_001244766
NM_001244767
NM_173468
|
MOB1B
|
MOB kinase activator 1B
|
|
chr2_+_24807345
|
0.140
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr3_+_88188261
|
0.138
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_-_185126153
|
0.137
|
NM_001202423
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr6_-_119670897
|
0.134
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr3_-_53381533
|
0.134
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr6_-_144385647
|
0.134
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr12_-_56652118
|
0.133
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr20_+_51588945
|
0.133
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr20_+_278200
|
0.132
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr3_-_24536176
|
0.132
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr6_+_73076431
|
0.132
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_+_149535508
|
0.131
|
NM_001099220
|
ZNF862
|
zinc finger protein 862
|
|
chr11_+_33279167
|
0.131
|
NM_001048200
NM_005734
|
HIPK3
|
homeodomain interacting protein kinase 3
|