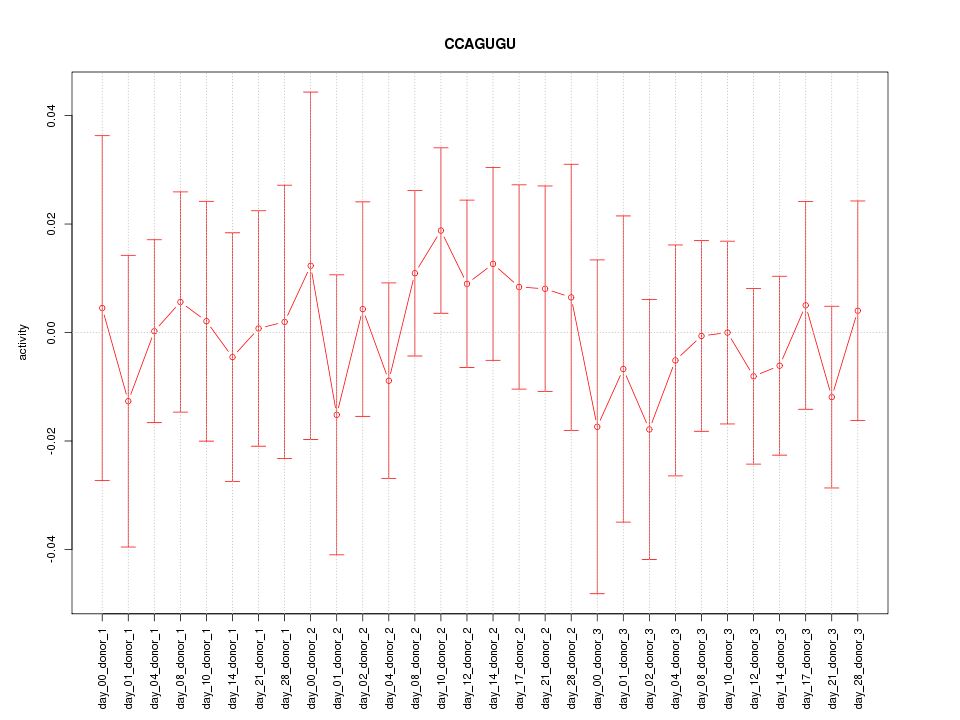
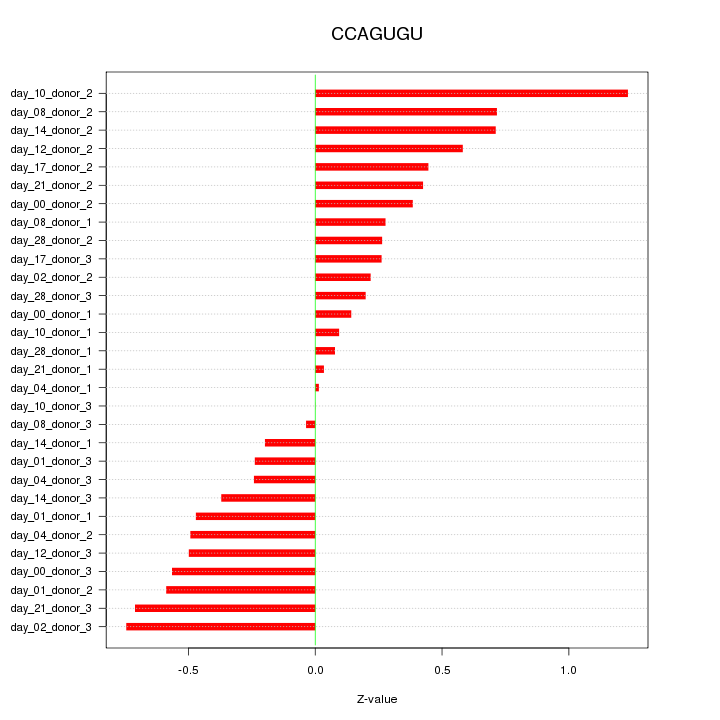
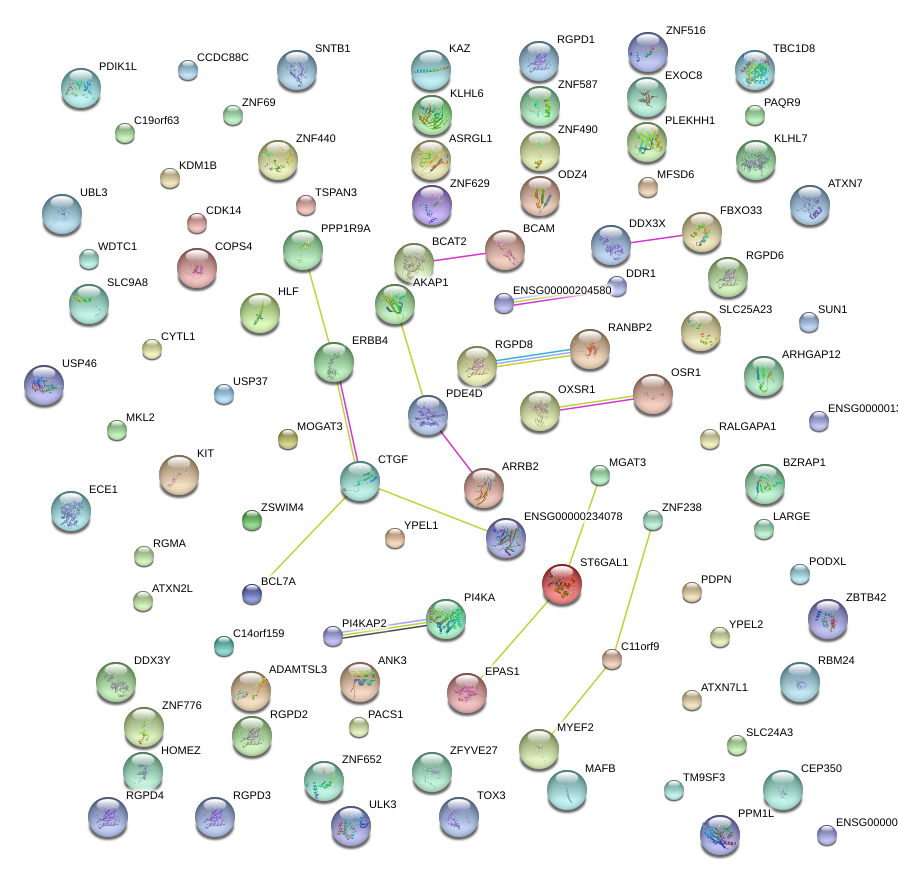
|
chr7_-_131241321
|
1.095
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr16_-_52580805
|
1.027
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr16_-_52581713
|
1.013
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr6_+_17281773
|
0.900
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
0.886
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282931
|
0.882
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_+_186739643
|
0.658
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr14_+_67999915
|
0.632
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr4_+_55523906
|
0.612
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr3_+_186648273
|
0.588
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr2_-_19558371
|
0.585
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr15_-_93616891
|
0.471
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr17_+_53342320
|
0.468
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr3_-_183273407
|
0.465
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr15_-_93617091
|
0.455
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93632161
|
0.452
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr8_-_121824287
|
0.452
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr15_-_93631751
|
0.450
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616358
|
0.441
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chrY_+_15016018
|
0.441
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr15_-_48470453
|
0.426
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr14_-_91884164
|
0.410
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr20_+_19193282
|
0.407
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr22_+_39853316
|
0.396
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr10_-_61900659
|
0.393
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_62149487
|
0.388
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_+_105266878
|
0.352
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr22_+_39883228
|
0.348
|
NM_001098270
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr10_-_62332666
|
0.338
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_13911966
|
0.312
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr6_+_18155535
|
0.299
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr1_+_13910251
|
0.298
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr10_-_62493282
|
0.295
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrY_+_15016698
|
0.291
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr19_+_45312294
|
0.286
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr17_-_56406120
|
0.272
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr1_+_244214552
|
0.259
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr20_+_48429249
|
0.254
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr15_+_84322809
|
0.252
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr18_-_74207145
|
0.252
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr2_-_219432955
|
0.250
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr2_-_213403280
|
0.245
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr7_+_94536926
|
0.243
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr11_+_61520000
|
0.238
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr7_+_872137
|
0.237
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr1_+_244216506
|
0.232
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr6_-_132272311
|
0.231
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr7_-_105319608
|
0.229
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr14_-_36278335
|
0.228
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr16_+_14164879
|
0.226
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr7_+_94539209
|
0.221
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr19_+_58258160
|
0.220
|
NM_173632
|
ZNF776
|
zinc finger protein 776
|
|
chr3_+_160473995
|
0.212
|
NM_139245
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L
|
|
chr1_+_27561006
|
0.209
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr17_-_47439326
|
0.207
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr19_-_6459735
|
0.204
|
NM_024103
|
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr14_+_91580967
|
0.200
|
NM_001102367
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr3_-_142682177
|
0.195
|
NM_198504
|
PAQR9
|
progestin and adipoQ receptor family member IX
|
|
chr14_-_39901619
|
0.192
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr22_-_21088954
|
0.190
|
NM_002650
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr19_+_11925077
|
0.188
|
NM_152357
|
ZNF440
|
zinc finger protein 440
|
|
chr11_+_62104773
|
0.184
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr1_+_179923870
|
0.182
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr17_+_55162530
|
0.182
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_+_191273012
|
0.181
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr15_-_75135524
|
0.176
|
NM_001099436
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr11_+_61522860
|
0.173
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr10_+_99498233
|
0.173
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr4_-_53525316
|
0.172
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr20_-_39317875
|
0.171
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr13_-_30424795
|
0.170
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr10_+_99496847
|
0.169
|
NM_001002262
NM_001174119
NM_001174120
NM_001174121
NM_001174122
NM_144588
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr6_+_30856462
|
0.165
|
NM_001202521
NM_001202522
NM_013994
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr16_-_30798206
|
0.163
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr22_-_34316325
|
0.161
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr22_-_22089975
|
0.161
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr10_-_98346793
|
0.160
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_231473478
|
0.160
|
NM_175876
|
EXOC8
|
exocyst complex component 8
|
|
chr17_+_55163077
|
0.155
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr7_+_90338711
|
0.154
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr2_-_101767714
|
0.152
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr2_+_109335906
|
0.150
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr5_-_59783885
|
0.149
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_21671905
|
0.149
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr19_-_12721493
|
0.148
|
NM_020714
|
ZNF490
|
zinc finger protein 490
|
|
chr11_-_79151694
|
0.147
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr3_+_63884074
|
0.146
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr6_+_30852756
|
0.145
|
NM_001202523
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chrX_+_49644469
|
0.143
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr1_-_21605984
|
0.143
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr6_+_30852326
|
0.141
|
NM_013993
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr19_+_50979656
|
0.140
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr1_+_26438267
|
0.139
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr12_+_122459791
|
0.139
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr10_-_32217762
|
0.139
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr6_+_30851860
|
0.139
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr17_+_4613780
|
0.137
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chr19_+_13906207
|
0.136
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr15_-_77363519
|
0.130
|
NM_001168412
NM_005724
NM_198902
|
TSPAN3
|
tetraspanin 3
|
|
chr10_+_69644938
|
0.130
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr17_-_16118789
|
0.129
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr22_-_21213099
|
0.128
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr6_-_119031230
|
0.127
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr22_-_46373007
|
0.124
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr22_+_31031672
|
0.122
|
NM_001001479
|
SLC35E4
|
solute carrier family 35, member E4
|
|
chr12_+_46123495
|
0.121
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr15_+_43809805
|
0.120
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr4_-_23891608
|
0.120
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr20_-_4795768
|
0.119
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr19_-_58662144
|
0.117
|
NM_024620
|
ZNF329
|
zinc finger protein 329
|
|
chr8_-_77913279
|
0.117
|
NM_001172086
NM_001172087
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr5_+_172483285
|
0.116
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_-_153895450
|
0.115
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr9_-_111882209
|
0.115
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr10_+_69644341
|
0.115
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr1_-_21616648
|
0.115
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr12_+_11802726
|
0.115
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr14_-_23388380
|
0.115
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr20_-_4804067
|
0.113
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr14_+_103243813
|
0.113
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr4_-_53522756
|
0.110
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr10_-_14816895
|
0.109
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr20_-_62436855
|
0.108
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr7_+_140774031
|
0.108
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr16_+_4382224
|
0.106
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr1_-_182558344
|
0.105
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr5_-_58571916
|
0.105
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_52345437
|
0.104
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr5_-_59189620
|
0.103
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_78531848
|
0.103
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr8_+_110346549
|
0.102
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr17_-_58603492
|
0.102
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr1_+_36348795
|
0.102
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr3_+_63953419
|
0.102
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr14_+_91580356
|
0.100
|
NM_001102366
NM_001102368
NM_001102369
NM_024952
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr12_+_93771607
|
0.100
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr2_-_242212244
|
0.100
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr7_-_105516923
|
0.099
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr5_-_693413
|
0.097
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr3_-_52719514
|
0.096
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr2_-_217560247
|
0.095
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr3_-_15106806
|
0.095
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr5_-_115910406
|
0.094
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr1_+_218518675
|
0.093
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr2_+_148602569
|
0.093
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr11_-_118661898
|
0.093
|
NM_004397
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr8_-_102217907
|
0.093
|
NM_001042510
NM_016096
|
ZNF706
|
zinc finger protein 706
|
|
chr11_+_121322848
|
0.093
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chrX_+_70315637
|
0.091
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr9_-_128003634
|
0.090
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr12_-_9102262
|
0.089
|
NM_001207024
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr20_+_11898536
|
0.089
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr8_-_102217868
|
0.088
|
NM_001042511
|
ZNF706
|
zinc finger protein 706
|
|
chr19_-_58400287
|
0.087
|
NM_001144989
|
ZNF814
|
zinc finger protein 814
|
|
chr8_-_77912296
|
0.087
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr8_+_98881277
|
0.087
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr8_-_71316006
|
0.087
|
NM_006540
|
NCOA2
|
nuclear receptor coactivator 2
|
|
chr11_-_102323353
|
0.086
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr19_+_49660997
|
0.085
|
NM_001195227
NM_017636
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4
|
|
chr1_-_19282773
|
0.084
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr2_+_63277191
|
0.082
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_-_21616888
|
0.081
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_+_36583369
|
0.081
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr17_+_6659155
|
0.080
|
NM_017523
NM_199139
|
XAF1
|
XIAP associated factor 1
|
|
chr19_+_12902295
|
0.080
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr3_-_189840225
|
0.079
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr1_+_172502291
|
0.079
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr3_+_38495778
|
0.079
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr5_-_58295758
|
0.078
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58334936
|
0.078
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_78740508
|
0.078
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr4_-_46996423
|
0.078
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr1_+_172502012
|
0.077
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr14_-_50698090
|
0.077
|
NM_006939
|
SOS2
|
son of sevenless homolog 2 (Drosophila)
|
|
chr17_-_27916602
|
0.076
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr22_-_46933066
|
0.076
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr5_+_139493689
|
0.075
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr2_+_63277936
|
0.075
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr8_+_42128797
|
0.074
|
NM_001190720
NM_001242778
NM_001556
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr3_+_88188261
|
0.074
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr3_-_113415313
|
0.073
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr17_-_44270255
|
0.073
|
NM_015443
|
KIAA1267
|
KIAA1267
|
|
chr11_-_62414096
|
0.073
|
NM_198334
NM_198335
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr3_+_180630054
|
0.072
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr12_+_69633316
|
0.072
|
NM_007007
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa
|
|
chr17_+_43224683
|
0.072
|
NM_006460
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr16_+_55512801
|
0.072
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr5_-_58652671
|
0.072
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_-_135863502
|
0.071
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr17_-_44270144
|
0.070
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr3_+_63850181
|
0.070
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr7_+_130794854
|
0.070
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr6_-_137113624
|
0.069
|
NM_005923
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr2_-_222436996
|
0.069
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr19_+_45349392
|
0.068
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr17_+_26369617
|
0.068
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr4_+_4387982
|
0.068
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr17_-_44302716
|
0.067
|
NM_001193465
|
KIAA1267
|
KIAA1267
|
|
chr17_-_6544246
|
0.067
|
NM_014804
|
KIAA0753
|
KIAA0753
|
|
chr20_-_4982133
|
0.067
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|