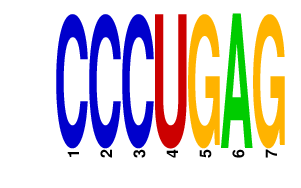
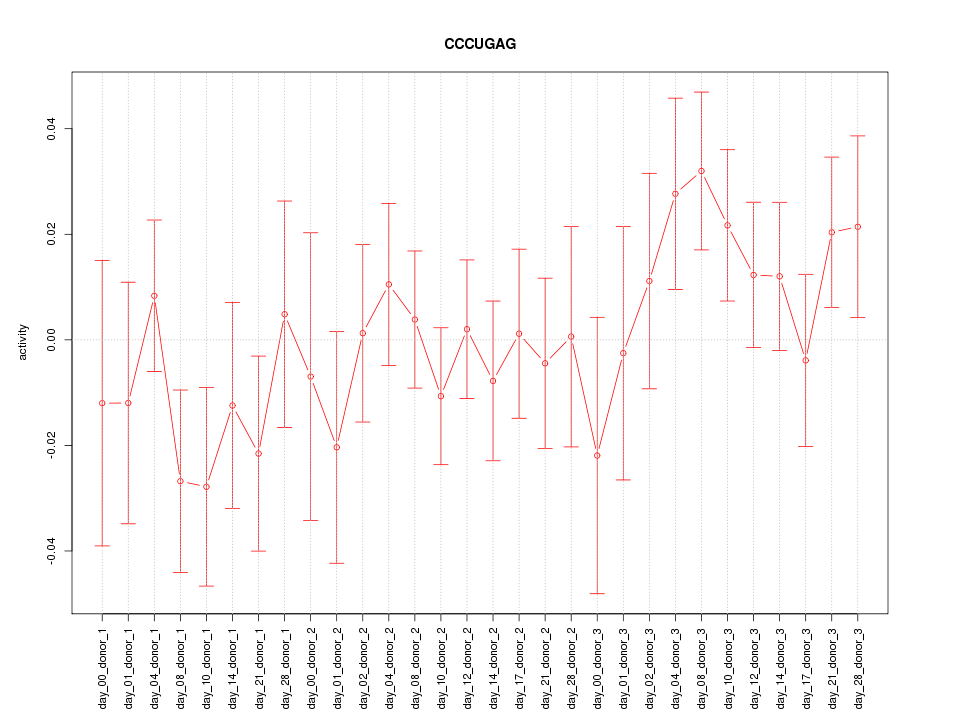
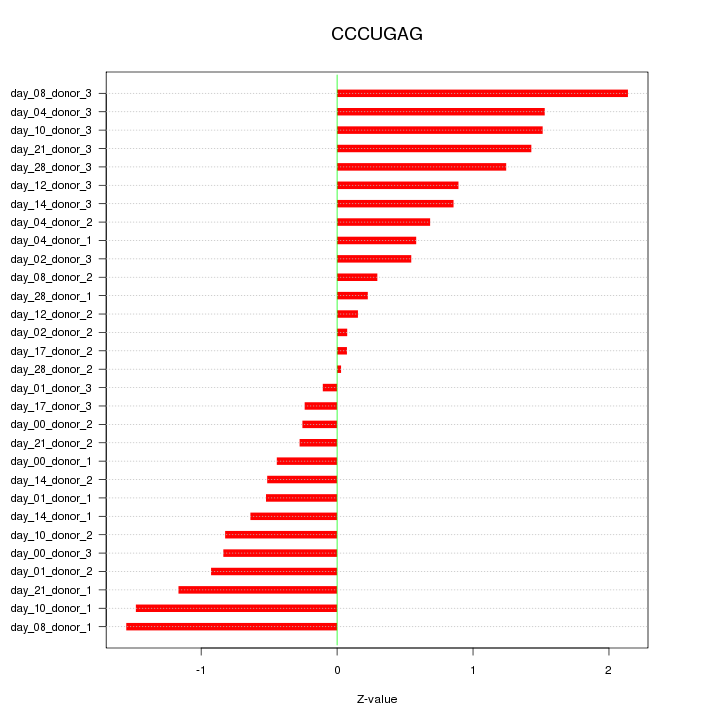
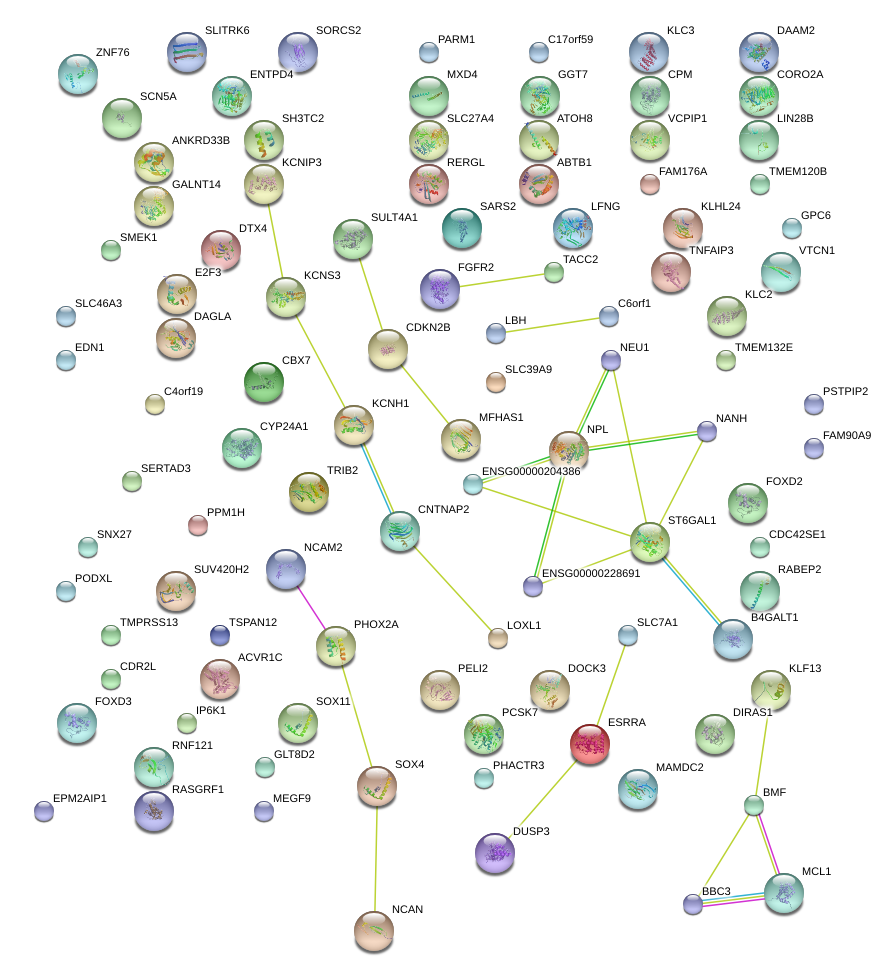
|
chr4_+_75858205
|
2.790
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr20_+_58179601
|
1.115
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58251715
|
1.097
|
NM_001199506
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58152563
|
1.071
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58296264
|
1.067
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_-_117102810
|
0.852
|
NM_004716
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr12_-_63328663
|
0.805
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr6_+_138188352
|
0.783
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_31361591
|
0.757
|
NM_001253826
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr4_+_7194317
|
0.726
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr1_-_117753476
|
0.724
|
NM_001253850
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr19_-_2721337
|
0.712
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr2_+_96012767
|
0.711
|
NM_001034914
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr5_-_148442606
|
0.701
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr2_+_85980600
|
0.683
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr19_+_55851220
|
0.647
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr11_+_66024746
|
0.644
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr9_-_33167355
|
0.637
|
NM_001497
|
B4GALT1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1
|
|
chr11_+_71639767
|
0.619
|
NM_018320
|
RNF121
|
ring finger protein 121
|
|
chr2_+_30454396
|
0.615
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr6_+_105404922
|
0.609
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr1_+_182758583
|
0.609
|
NM_001200050
NM_001200051
NM_001200052
NM_001200056
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr5_+_10564431
|
0.606
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr15_-_40401041
|
0.604
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr3_+_127391768
|
0.588
|
NM_032548
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr3_+_183353355
|
0.581
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr20_-_52790511
|
0.577
|
NM_000782
NM_001128915
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1
|
|
chr17_-_41856180
|
0.568
|
NM_004090
|
DUSP3
|
dual specificity phosphatase 3
|
|
chr3_+_186648273
|
0.561
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr3_+_50712671
|
0.559
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr13_-_86373328
|
0.557
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr12_-_69357019
|
0.547
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr13_-_29292961
|
0.543
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr12_-_69326946
|
0.543
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr3_+_186739643
|
0.542
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr14_-_91976630
|
0.530
|
NM_032560
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium)
|
|
chr1_-_151032082
|
0.523
|
NM_001038707
NM_020239
|
CDC42SE1
|
CDC42 small effector 1
|
|
chr22_-_44258210
|
0.511
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr2_+_95963026
|
0.508
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr2_+_18059923
|
0.499
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr15_-_40398573
|
0.489
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr9_-_100935173
|
0.488
|
NM_003389
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr8_-_8750739
|
0.477
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr7_-_131241321
|
0.477
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr17_+_72983723
|
0.476
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr17_-_8093492
|
0.474
|
NM_017622
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr9_-_123476578
|
0.461
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr19_-_47734215
|
0.458
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr7_+_145813452
|
0.454
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr1_+_47901688
|
0.442
|
NM_004474
|
FOXD2
|
forkhead box D2
|
|
chr15_+_74218786
|
0.442
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr8_-_23315176
|
0.434
|
NM_001128930
NM_004901
|
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4
|
|
chr9_+_72658496
|
0.431
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr12_+_122150638
|
0.427
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr20_-_33460558
|
0.427
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr9_+_131102786
|
0.422
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr3_-_38691118
|
0.413
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr2_+_12856813
|
0.413
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr13_-_30169795
|
0.411
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr2_-_75788079
|
0.405
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr6_+_20402040
|
0.399
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr9_-_100954883
|
0.398
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr14_+_56584854
|
0.395
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr19_-_40950238
|
0.394
|
NM_203344
|
SERTAD3
|
SERTA domain containing 3
|
|
chr11_+_64073040
|
0.394
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr6_+_35227478
|
0.392
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr15_-_40398286
|
0.389
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr13_+_93879011
|
0.382
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr7_+_2557496
|
0.376
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr14_+_69865380
|
0.374
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr2_-_158454025
|
0.371
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr3_-_49823626
|
0.371
|
NM_001242829
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr1_+_151584514
|
0.368
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr3_-_49823972
|
0.367
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr21_+_22370622
|
0.364
|
NM_004540
|
NCAM2
|
neural cell adhesion molecule 2
|
|
chr11_+_66025070
|
0.363
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr1_-_211307314
|
0.360
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr2_+_5832778
|
0.357
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr19_-_47735858
|
0.355
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr19_+_19322763
|
0.354
|
NM_004386
|
NCAN
|
neurocan
|
|
chr7_-_120498128
|
0.354
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr22_-_39548448
|
0.352
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr4_-_2263706
|
0.352
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr10_+_123923104
|
0.351
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr15_-_79298001
|
0.351
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr15_+_31619043
|
0.350
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr17_+_32907767
|
0.349
|
NM_207313
|
TMEM132E
|
transmembrane protein 132E
|
|
chr16_-_28936454
|
0.348
|
NM_024816
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr6_+_12290528
|
0.346
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr1_-_150552127
|
0.345
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr10_-_123353771
|
0.344
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr4_+_37585854
|
0.343
|
NM_018302
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr6_+_39760148
|
0.343
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr7_+_2552162
|
0.343
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr4_+_37455551
|
0.340
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr18_-_43652153
|
0.334
|
NM_024430
|
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2
|
|
chr9_-_22009270
|
0.334
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr11_-_117800067
|
0.333
|
NM_001077263
NM_001206789
NM_001206790
NM_001244995
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr8_-_67579446
|
0.332
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr6_-_31830593
|
0.331
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr2_-_75796847
|
0.330
|
NM_032181
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr11_+_58939539
|
0.328
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr11_+_61447816
|
0.328
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr3_-_118753665
|
0.327
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr17_-_40540359
|
0.326
|
NM_213662
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr2_+_191745546
|
0.326
|
NM_014905
|
GLS
|
glutaminase
|
|
chr11_-_77899640
|
0.322
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr12_+_69633316
|
0.322
|
NM_007007
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa
|
|
chr2_-_158485393
|
0.317
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr19_-_40948504
|
0.316
|
NM_013368
|
SERTAD3
|
SERTA domain containing 3
|
|
chr2_+_46769821
|
0.313
|
NM_012249
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr6_-_43596919
|
0.310
|
NM_019096
|
GTPBP2
|
GTP binding protein 2
|
|
chr6_+_39760772
|
0.308
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr8_-_17579729
|
0.307
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr19_+_45394459
|
0.305
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr17_+_41177247
|
0.304
|
NM_005440
|
RND2
|
Rho family GTPase 2
|
|
chr1_-_6321034
|
0.302
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr13_-_53024730
|
0.301
|
NM_016075
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae)
|
|
chr20_-_32262254
|
0.300
|
NM_031231
NM_031232
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr13_-_73356070
|
0.299
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr19_-_18632886
|
0.298
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr2_+_150186498
|
0.297
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr9_+_12774988
|
0.297
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr10_-_71906452
|
0.294
|
NM_001040273
NM_173555
|
TYSND1
|
trypsin domain containing 1
|
|
chr17_+_34058678
|
0.290
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr4_-_25032306
|
0.290
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr15_-_61521478
|
0.285
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_+_1411089
|
0.285
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr2_-_99347588
|
0.278
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr9_+_139981414
|
0.277
|
NM_016219
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1
|
|
chr19_-_15560761
|
0.276
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr3_+_126707380
|
0.273
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr17_-_40540491
|
0.272
|
NM_003150
NM_139276
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr19_-_7293876
|
0.272
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr14_-_73453651
|
0.272
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr16_+_30772932
|
0.270
|
NM_001207033
NM_001207034
NM_014771
|
RNF40
|
ring finger protein 40
|
|
chr14_-_23755308
|
0.270
|
NM_020834
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr6_+_148663728
|
0.270
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr20_+_54933982
|
0.269
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr19_-_49865638
|
0.265
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr16_+_66400524
|
0.265
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr11_+_45825622
|
0.264
|
NM_001145265
NM_001145266
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr3_-_13461802
|
0.262
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr20_+_62795826
|
0.262
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr15_+_45315301
|
0.260
|
NM_003104
|
SORD
|
sorbitol dehydrogenase
|
|
chr14_+_70078309
|
0.257
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr7_-_6746541
|
0.256
|
NM_006956
NM_016265
|
ZNF12
|
zinc finger protein 12
|
|
chr3_-_118864881
|
0.254
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr15_-_43662107
|
0.253
|
NM_152455
|
ZSCAN29
|
zinc finger and SCAN domain containing 29
|
|
chr10_-_123356158
|
0.253
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr10_+_123748688
|
0.251
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr19_+_7968764
|
0.250
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr11_-_65381641
|
0.249
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr5_+_139944411
|
0.248
|
NM_080670
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr2_+_7057471
|
0.248
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr17_-_61777478
|
0.247
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr1_-_38230804
|
0.246
|
NM_001099439
NM_173641
|
EPHA10
|
EPH receptor A10
|
|
chr2_+_150187028
|
0.246
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr12_-_120806893
|
0.245
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr17_+_21279667
|
0.244
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr12_-_9102262
|
0.243
|
NM_001207024
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr11_+_45826640
|
0.243
|
NM_018389
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr12_-_57030083
|
0.241
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr5_+_133861685
|
0.239
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr17_-_62050200
|
0.238
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr20_+_42086468
|
0.237
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr17_-_66287256
|
0.236
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr17_-_37844266
|
0.235
|
NM_033419
|
PGAP3
|
post-GPI attachment to proteins 3
|
|
chr16_+_57406398
|
0.230
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr6_-_117923692
|
0.229
|
NM_001017408
NM_020399
|
GOPC
|
golgi-associated PDZ and coiled-coil motif containing
|
|
chr17_+_42836546
|
0.228
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr22_-_37098620
|
0.227
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr6_+_132129154
|
0.227
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr5_+_78531848
|
0.227
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr1_-_151319717
|
0.227
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr16_+_71929432
|
0.225
|
NM_014761
|
IST1
|
increased sodium tolerance 1 homolog (yeast)
|
|
chr16_+_8891669
|
0.223
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr16_-_19729417
|
0.223
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr1_+_57110745
|
0.221
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr20_+_37434294
|
0.220
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr11_-_66335958
|
0.220
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr17_-_40333190
|
0.220
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr12_+_11802726
|
0.219
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chrX_-_53310748
|
0.217
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr17_+_53828440
|
0.217
|
NM_001102402
|
PCTP
|
phosphatidylcholine transfer protein
|
|
chrX_-_128977719
|
0.214
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr10_-_5708542
|
0.213
|
NM_024701
|
ASB13
|
ankyrin repeat and SOCS box containing 13
|
|
chr9_-_130829598
|
0.212
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr3_-_48672925
|
0.212
|
NM_022911
NM_134263
NM_134426
|
SLC26A6
|
solute carrier family 26, member 6
|
|
chr15_-_60919642
|
0.211
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_-_156470506
|
0.210
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr7_+_75831194
|
0.208
|
NM_001110199
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr5_+_156887026
|
0.206
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr1_+_214161859
|
0.206
|
NM_002763
|
PROX1
|
prospero homeobox 1
|
|
chr15_+_73344824
|
0.205
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr7_-_139477437
|
0.205
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr5_-_133968463
|
0.204
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr1_-_161168816
|
0.204
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr8_-_17580024
|
0.204
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_8806825
|
0.203
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|