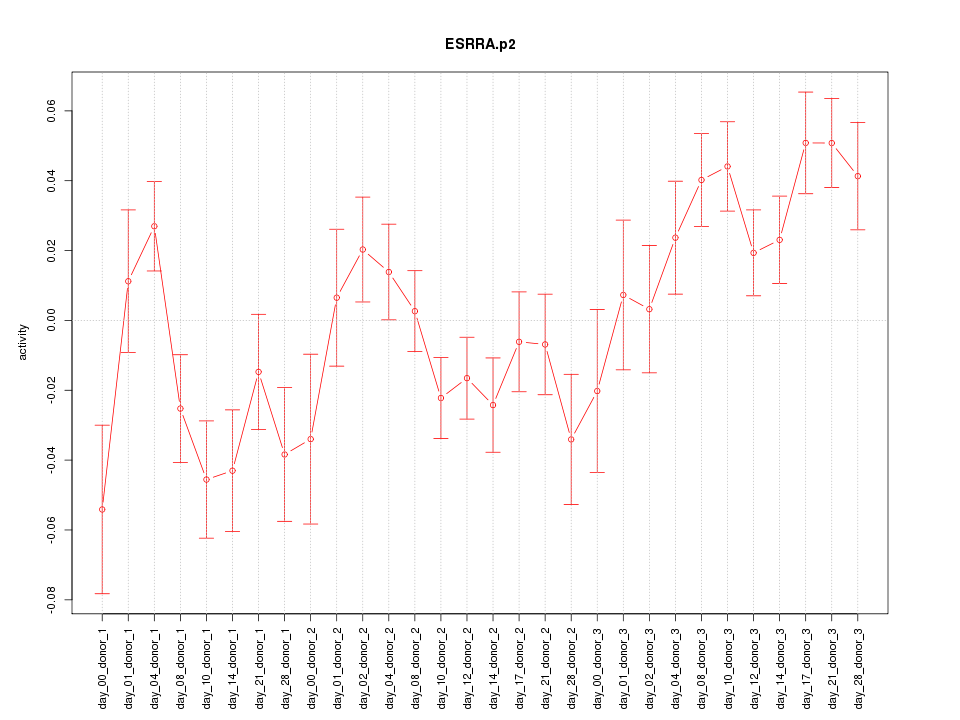
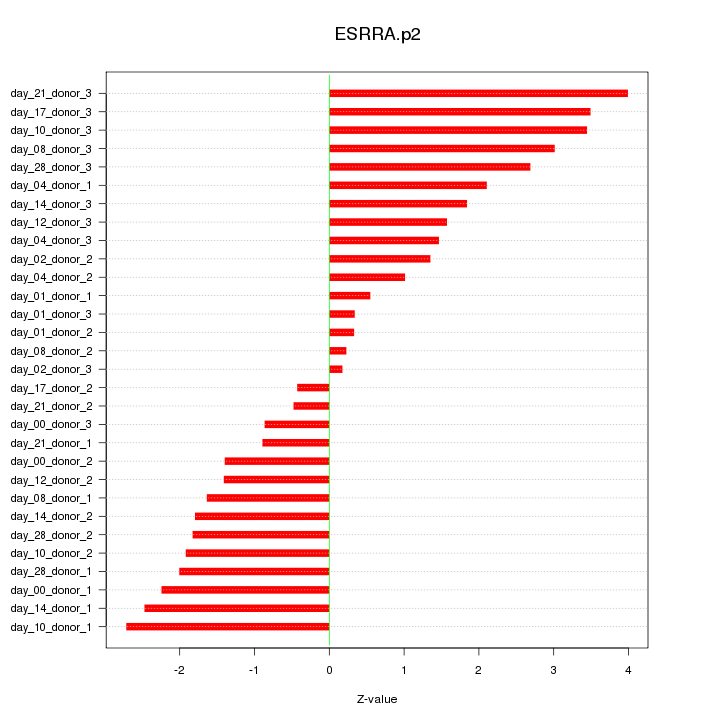
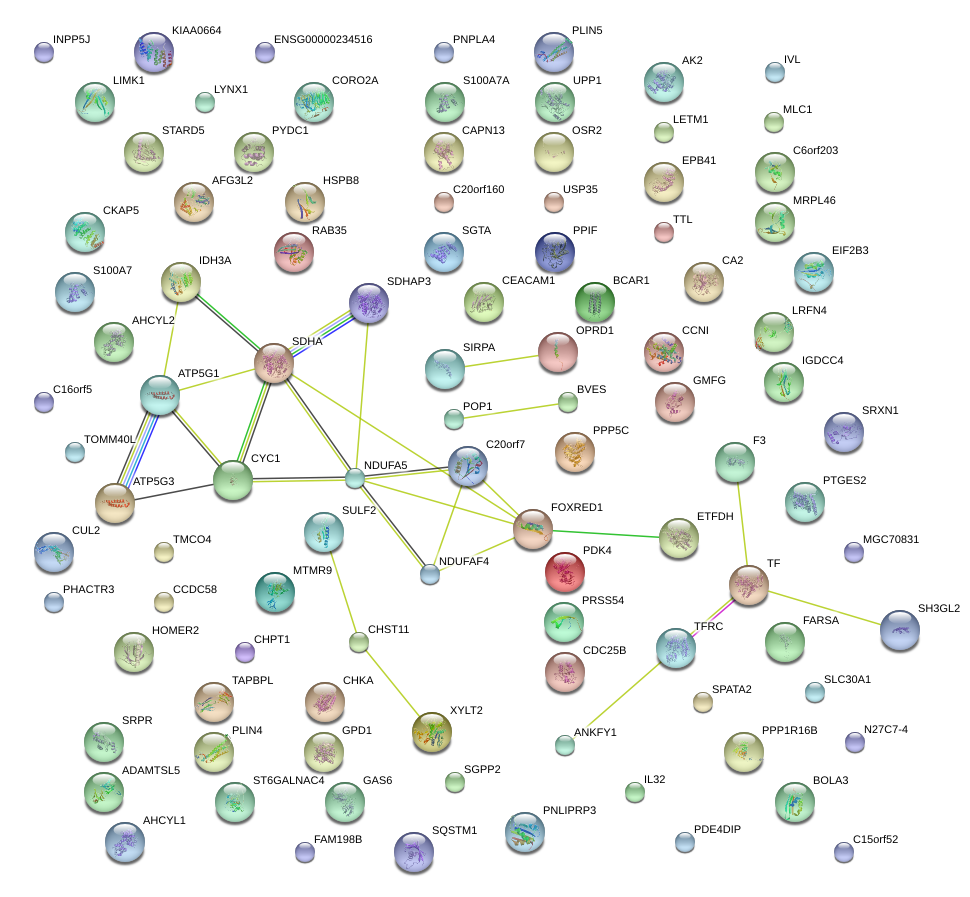
|
chr10_+_118187381
|
6.358
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr20_+_58179601
|
6.016
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr1_+_152881022
|
4.272
|
NM_005547
|
IVL
|
involucrin
|
|
chr5_+_218447
|
3.632
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr5_+_218355
|
3.268
|
NM_004168
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr2_-_31030271
|
3.192
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr10_+_81107242
|
3.062
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr3_+_133465250
|
2.951
|
|
TF
|
transferrin
|
|
chr3_+_133465098
|
2.883
|
|
TF
|
transferrin
|
|
chr3_+_133465204
|
2.803
|
|
TF
|
transferrin
|
|
chr3_+_133464883
|
2.685
|
NM_001063
|
TF
|
transferrin
|
|
chr1_-_144994908
|
2.675
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chrX_-_7895764
|
2.626
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr5_+_218441
|
2.595
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr12_+_102091399
|
2.432
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr12_-_120554558
|
2.358
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr10_+_81107259
|
2.330
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr11_+_66624501
|
2.118
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr20_-_46415277
|
2.070
|
NM_001161841
NM_198596
|
SULF2
|
sulfatase 2
|
|
chr13_-_114566951
|
2.045
|
|
GAS6
|
growth arrest-specific 6
|
|
chr13_-_114567034
|
2.041
|
|
GAS6
|
growth arrest-specific 6
|
|
chr13_-_114567040
|
2.029
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr1_-_153433104
|
2.024
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr19_-_1512997
|
2.015
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr12_-_120554551
|
2.009
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr13_-_114567011
|
1.971
|
|
GAS6
|
growth arrest-specific 6
|
|
chr19_-_1513187
|
1.921
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr12_-_120554453
|
1.899
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr7_+_129008044
|
1.898
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr15_-_81616439
|
1.893
|
NM_181900
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5
|
|
chr11_+_126138934
|
1.892
|
NM_017547
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr11_+_126139049
|
1.845
|
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr11_+_126139022
|
1.832
|
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr19_-_39826625
|
1.820
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr20_+_37434294
|
1.758
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr8_-_143858436
|
1.686
|
NM_177476
NM_177477
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr8_+_99956630
|
1.660
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr9_+_17578952
|
1.658
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr20_+_1876006
|
1.635
|
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr7_+_73498318
|
1.614
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr18_-_12377168
|
1.579
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr16_-_4588379
|
1.577
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr2_-_176046120
|
1.568
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr19_-_43031343
|
1.525
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr9_-_130889993
|
1.522
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr22_+_31518923
|
1.521
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr22_-_24110054
|
1.509
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr7_+_129007942
|
1.506
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_+_81107228
|
1.492
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr1_-_144994839
|
1.482
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr20_+_1875825
|
1.475
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr10_+_81107219
|
1.435
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr20_+_3776905
|
1.432
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr12_+_6561176
|
1.431
|
NM_018009
|
TAPBPL
|
TAP binding protein-like
|
|
chr7_-_95225802
|
1.403
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr20_+_3776385
|
1.390
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr6_-_97345747
|
1.361
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr1_-_45452215
|
1.359
|
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr9_-_130890007
|
1.357
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr9_-_130889965
|
1.352
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr1_+_161195832
|
1.342
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr12_+_105153258
|
1.340
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr16_-_58328879
|
1.326
|
NM_001080492
|
PRSS54
|
protease, serine, 54
|
|
chr8_+_86376130
|
1.325
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr9_-_130890026
|
1.314
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr6_-_97345676
|
1.298
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr19_-_39826676
|
1.288
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr16_-_75285383
|
1.265
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr20_-_48532036
|
1.250
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr15_-_65715385
|
1.248
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr11_+_77899857
|
1.231
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr1_+_29213619
|
1.230
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr8_+_99129488
|
1.226
|
NM_001145860
NM_001145861
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr19_-_4535202
|
1.226
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr4_+_159593233
|
1.169
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr2_+_113239733
|
1.160
|
NM_153712
|
TTL
|
tubulin tyrosine ligase
|
|
chr12_+_119616594
|
1.155
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr9_-_130679210
|
1.121
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr1_-_211751953
|
1.118
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr3_-_195808980
|
1.118
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr6_-_97345760
|
1.106
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr19_-_13044459
|
1.106
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr17_+_48423382
|
1.084
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr15_+_78441737
|
1.067
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr19_-_4517715
|
1.065
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr17_+_46970128
|
1.059
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr3_-_122102057
|
1.053
|
NM_001017928
|
CCDC58
|
coiled-coil domain containing 58
|
|
chr19_+_46850245
|
1.043
|
NM_001204284
NM_006247
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr12_-_120554557
|
1.042
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr20_+_13765645
|
1.029
|
NM_001039375
NM_024120
|
C20orf7
|
chromosome 20 open reading frame 7
|
|
chr8_+_11141983
|
1.024
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr15_-_40630272
|
1.018
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr1_-_33502484
|
1.009
|
NM_001199199
NM_001625
NM_013411
|
AK2
|
adenylate kinase 2
|
|
chr5_+_179233351
|
1.005
|
NM_001142298
|
SQSTM1
|
sequestosome 1
|
|
chr16_+_3115312
|
1.000
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr4_-_1857677
|
0.992
|
NM_012318
|
LETM1
|
leucine zipper-EF-hand containing transmembrane protein 1
|
|
chr2_+_223289212
|
0.989
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr20_+_30598244
|
0.979
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr1_-_20126368
|
0.969
|
NM_181719
|
TMCO4
|
transmembrane and coiled-coil domains 4
|
|
chr17_-_2614926
|
0.967
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr7_+_48128939
|
0.949
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr15_-_83621350
|
0.948
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr7_+_48128738
|
0.946
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr7_-_123197824
|
0.942
|
NM_005000
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa
|
|
chr15_+_78441678
|
0.936
|
NM_005530
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr4_-_159094163
|
0.932
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_+_29138653
|
0.930
|
NM_000911
|
OPRD1
|
opioid receptor, delta 1
|
|
chr6_+_107349327
|
0.924
|
NM_001142468
NM_001142470
NM_016487
|
C6orf203
|
chromosome 6 open reading frame 203
|
|
chr19_-_2783225
|
0.919
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr18_-_12377248
|
0.918
|
NM_006796
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr20_-_48531986
|
0.909
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr2_-_74374998
|
0.896
|
NM_001035505
NM_212552
|
BOLA3
|
bolA homolog 3 (E. coli)
|
|
chr22_-_50523853
|
0.896
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr22_-_50523780
|
0.895
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr9_-_100954883
|
0.895
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr5_+_218428
|
0.887
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr10_-_35363167
|
0.880
|
NM_001198778
NM_001198779
|
CUL2
|
cullin 2
|
|
chr15_+_78441733
|
0.877
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr8_+_145149985
|
0.877
|
|
CYC1
|
cytochrome c-1
|
|
chr11_-_126138756
|
0.876
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr17_-_4167028
|
0.875
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr20_-_633808
|
0.870
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr11_-_46867788
|
0.855
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr11_-_126138778
|
0.854
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr11_-_67888857
|
0.854
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr22_-_50524357
|
0.849
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr11_-_67888670
|
0.848
|
|
CHKA
|
choline kinase alpha
|
|
chr8_+_145149936
|
0.848
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr11_-_126138789
|
0.846
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr22_-_50523737
|
0.842
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr15_-_89010607
|
0.842
|
NM_022163
|
MRPL46
|
mitochondrial ribosomal protein L46
|
|
chr11_+_66059364
|
0.840
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr16_+_8814567
|
0.837
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr19_+_46850344
|
0.825
|
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr6_-_33281790
|
0.822
|
|
TAPBP
|
TAP binding protein (tapasin)
|
|
chr3_-_42845911
|
0.817
|
NM_001099668
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr4_-_170947428
|
0.794
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr1_-_241683011
|
0.793
|
|
FH
|
fumarate hydratase
|
|
chr3_-_183735617
|
0.791
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr19_+_45394642
|
0.790
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr10_-_50970321
|
0.784
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr2_+_113239812
|
0.783
|
|
TTL
|
tubulin tyrosine ligase
|
|
chr20_-_633844
|
0.768
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr9_-_37904331
|
0.763
|
NM_033412
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr17_-_72855884
|
0.753
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr1_+_207226594
|
0.751
|
NM_001018053
NM_006212
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr11_-_126138824
|
0.747
|
NM_001177842
NM_003139
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chrX_-_129299680
|
0.747
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chrX_-_129299803
|
0.735
|
NM_001130847
NM_004208
NM_145812
NM_145813
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr5_+_149569519
|
0.733
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr19_+_40146533
|
0.731
|
NM_001190441
|
LGALS16
|
lectin, galactoside-binding, soluble, 16
|
|
chr3_-_179322410
|
0.727
|
NM_020409
NM_177988
|
MRPL47
|
mitochondrial ribosomal protein L47
|
|
chr19_-_5622794
|
0.724
|
NM_014649
|
SAFB2
|
scaffold attachment factor B2
|
|
chr1_-_31712733
|
0.721
|
NM_024522
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chr14_+_57735605
|
0.718
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr7_+_2443164
|
0.718
|
NM_001243794
NM_001243795
NM_018641
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12
|
|
chrX_-_129299629
|
0.713
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr3_-_14166370
|
0.713
|
NM_001098502
NM_144636
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr11_+_66059335
|
0.711
|
|
TMEM151A
|
transmembrane protein 151A
|
|
chr7_-_1199785
|
0.707
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr1_+_205538164
|
0.706
|
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr16_+_4674819
|
0.699
|
NM_001142289
NM_001142290
NM_001142291
NM_015246
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr10_-_91404901
|
0.696
|
|
PANK1
|
pantothenate kinase 1
|
|
chr11_+_65383631
|
0.694
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|
|
chr3_-_195808958
|
0.693
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr4_-_15939949
|
0.689
|
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr5_-_137911317
|
0.687
|
NM_004134
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr7_-_1199822
|
0.684
|
NM_182491
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr3_-_195808934
|
0.681
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr16_+_4674829
|
0.672
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr16_-_2581358
|
0.665
|
NM_001048212
|
CEMP1
|
cementum protein 1
|
|
chr22_-_25758523
|
0.665
|
NM_182492
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like
|
|
chr1_-_27481400
|
0.664
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr1_-_1690080
|
0.661
|
NM_001198995
|
NADK
|
NAD kinase
|
|
chr5_-_137911005
|
0.660
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr8_+_110346549
|
0.658
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr10_+_96443250
|
0.652
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr8_-_22550057
|
0.651
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr5_+_175084984
|
0.650
|
NM_001131055
|
HRH2
|
histamine receptor H2
|
|
chr16_-_75150664
|
0.636
|
NM_153486
NM_194436
|
LDHD
|
lactate dehydrogenase D
|
|
chr4_-_7069755
|
0.631
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr13_-_114526166
|
0.626
|
|
GAS6
|
growth arrest-specific 6
|
|
chr8_-_145550300
|
0.626
|
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr20_-_55100980
|
0.625
|
NM_080615
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7
|
|
chr3_-_14166228
|
0.620
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr9_+_132251403
|
0.618
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr12_-_120554612
|
0.618
|
NM_001167606
NM_006861
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr16_+_11038314
|
0.616
|
NM_001243403
NM_015226
|
CLEC16A
|
C-type lectin domain family 16, member A
|
|
chr1_-_211752052
|
0.615
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr13_+_99058147
|
0.613
|
|
|
|
|
chr22_-_29784568
|
0.612
|
NM_001127
NM_001166019
NM_145730
|
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit
|
|
chr2_+_198365012
|
0.611
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr19_+_39971496
|
0.610
|
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr11_-_68518910
|
0.608
|
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr14_-_24911658
|
0.607
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr19_+_10654591
|
0.607
|
NM_032885
|
ATG4D
|
ATG4 autophagy related 4 homolog D (S. cerevisiae)
|
|
chr7_-_1199813
|
0.606
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr19_-_40931880
|
0.600
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr3_-_113464991
|
0.600
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr1_-_19217384
|
0.598
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|