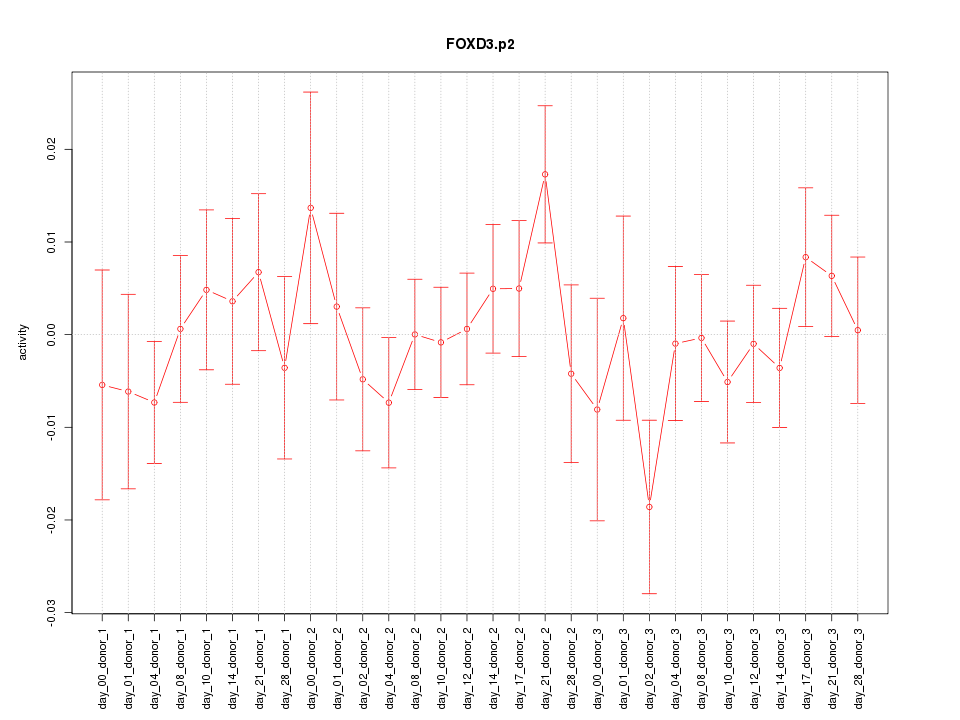
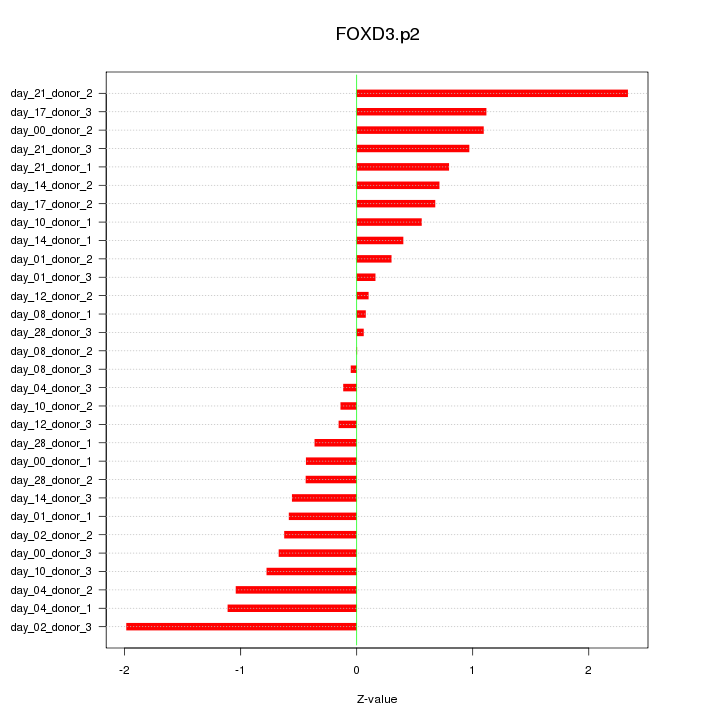
|
chr15_-_55800431
|
4.305
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|
|
chr20_+_56725982
|
4.090
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr12_+_49297892
|
4.044
|
NM_033124
|
CCDC65
|
coiled-coil domain containing 65
|
|
chr19_-_14992166
|
3.355
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chr9_-_34397786
|
2.615
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr12_-_122107559
|
2.597
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr9_+_105757592
|
2.474
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chrX_+_119029799
|
2.459
|
NM_001008534
NM_001008535
NM_178813
|
AKAP14
|
A kinase (PRKA) anchor protein 14
|
|
chr19_+_13134782
|
2.357
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr2_-_135782246
|
1.986
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr5_-_41213513
|
1.927
|
NM_000065
|
C6
|
complement component 6
|
|
chrX_-_44202834
|
1.792
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr4_+_186350543
|
1.770
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr4_-_16085593
|
1.711
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr3_-_183273407
|
1.696
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr4_+_72102249
|
1.688
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_+_159614373
|
1.673
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr7_-_16921596
|
1.578
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr5_+_140247767
|
1.519
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr13_+_43355685
|
1.439
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr5_+_94727047
|
1.374
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr18_+_61370193
|
1.356
|
NM_080475
|
SERPINB11
|
serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene)
|
|
chr13_-_86373328
|
1.285
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr11_+_73661363
|
1.277
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr11_+_6897855
|
1.259
|
NM_207186
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr15_+_78558522
|
1.244
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr9_-_4300028
|
1.213
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr4_-_70518964
|
1.204
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr18_-_24722757
|
1.199
|
NM_001243848
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr6_+_131958441
|
1.198
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr21_-_35884503
|
1.193
|
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr21_-_43735705
|
1.155
|
NM_003226
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr3_+_151488214
|
1.145
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr4_-_16085666
|
1.134
|
|
PROM1
|
prominin 1
|
|
chr5_-_110062365
|
1.114
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr9_-_117156684
|
1.097
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr5_-_35938736
|
1.089
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr2_+_170335948
|
1.071
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chr5_+_10441973
|
1.034
|
NM_001201466
NM_031916
|
ROPN1L
|
rhophilin associated tail protein 1-like
|
|
chr3_+_171561138
|
1.026
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr11_+_63137260
|
1.018
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr9_-_75567915
|
1.014
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr3_-_167098070
|
1.000
|
NM_001199202
NM_001199201
NM_024687
|
ZBBX
|
zinc finger, B-box domain containing
|
|
chr11_+_86085777
|
0.976
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr16_-_53737713
|
0.973
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr20_+_43990576
|
0.969
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr4_-_16077740
|
0.965
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr2_-_219031646
|
0.938
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr3_-_120401401
|
0.907
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr4_-_123377471
|
0.896
|
NM_000586
|
IL2
|
interleukin 2
|
|
chr9_-_75568232
|
0.890
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr2_+_61372206
|
0.889
|
NM_001143960
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr12_-_52779362
|
0.879
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr3_+_109128836
|
0.869
|
NM_001145553
|
FLJ25363
|
uncharacterized LOC401082
|
|
chr1_-_223853359
|
0.856
|
|
CAPN8
|
calpain 8
|
|
chr20_+_31870940
|
0.856
|
NM_033197
|
BPIFB1
|
BPI fold containing family B, member 1
|
|
chr3_+_132316080
|
0.855
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr1_-_151345156
|
0.819
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr16_+_67381252
|
0.819
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr11_-_102745666
|
0.798
|
NM_002426
|
MMP12
|
matrix metallopeptidase 12 (macrophage elastase)
|
|
chr19_-_55677919
|
0.797
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr19_+_14491920
|
0.783
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr18_-_53089722
|
0.779
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr4_+_70861647
|
0.775
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr19_+_41620336
|
0.767
|
NM_000774
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1
|
|
chr1_-_223853400
|
0.764
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chr9_+_127054246
|
0.762
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr12_-_71551778
|
0.753
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chr10_-_28287790
|
0.745
|
NM_018076
|
ARMC4
|
armadillo repeat containing 4
|
|
chr12_-_25801487
|
0.740
|
NM_001145727
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr3_-_114343052
|
0.740
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_9479943
|
0.738
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chr3_-_57530067
|
0.732
|
NM_178504
NM_198564
|
DNAH12
|
dynein, axonemal, heavy chain 12
|
|
chr9_+_102677457
|
0.731
|
|
STX17
|
syntaxin 17
|
|
chr1_-_57045214
|
0.729
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_+_93698959
|
0.720
|
NM_001174150
NM_001174151
NM_144996
NM_182896
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
chr2_+_132285469
|
0.719
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr14_-_61748193
|
0.714
|
|
|
|
|
chr6_-_52774460
|
0.700
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr2_-_128100673
|
0.695
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr2_-_217560247
|
0.694
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr5_+_75699003
|
0.689
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr7_+_89874487
|
0.686
|
NM_001039706
NM_001160138
|
C7orf63
|
chromosome 7 open reading frame 63
|
|
chr16_-_23724820
|
0.686
|
NM_033266
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2
|
|
chr1_+_244214576
|
0.682
|
|
ZNF238
|
zinc finger protein 238
|
|
chr13_+_50589389
|
0.677
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr5_+_140220811
|
0.676
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_140602914
|
0.673
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr11_+_73358593
|
0.672
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr1_-_57045235
|
0.669
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr4_+_154178525
|
0.668
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr7_+_142829169
|
0.665
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr14_-_81405883
|
0.662
|
NM_152446
|
CEP128
|
centrosomal protein 128kDa
|
|
chr10_-_32667543
|
0.648
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr16_-_28620648
|
0.644
|
NM_177530
NM_177534
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr17_-_5138049
|
0.638
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr13_-_36429798
|
0.636
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr21_-_43735462
|
0.629
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr12_+_9980076
|
0.617
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr12_-_25348031
|
0.616
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr1_-_170038055
|
0.612
|
NM_001204517
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr16_+_78056411
|
0.609
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr9_+_139927515
|
0.605
|
|
C9orf139
|
chromosome 9 open reading frame 139
|
|
chr1_+_244214552
|
0.599
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr13_+_24734788
|
0.598
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr13_-_41593405
|
0.595
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr13_-_41593491
|
0.593
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr6_+_43612766
|
0.590
|
NM_001193341
NM_152732
|
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas)
|
|
chr6_+_150690027
|
0.588
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr17_+_26646120
|
0.586
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chrX_-_24045302
|
0.585
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr12_+_20963637
|
0.578
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr19_+_15052298
|
0.571
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chr14_-_23288912
|
0.570
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr7_+_105603656
|
0.567
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr10_-_99161013
|
0.565
|
NM_001145114
NM_015179
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr2_-_175712276
|
0.565
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr3_-_120400957
|
0.564
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr7_+_114055048
|
0.564
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr20_+_9288446
|
0.564
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr6_-_154651214
|
0.557
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr8_-_116680208
|
0.557
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr1_+_244216506
|
0.556
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_205538164
|
0.556
|
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr10_-_116286570
|
0.552
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_36549714
|
0.552
|
|
TEKT2
|
tektin 2 (testicular)
|
|
chr18_-_47814691
|
0.551
|
NM_001101654
NM_014593
|
CXXC1
|
CXXC finger protein 1
|
|
chr5_-_149792276
|
0.550
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr15_-_66648991
|
0.547
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr20_+_19870209
|
0.545
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr9_-_14086901
|
0.544
|
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_167371288
|
0.544
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr22_-_43036606
|
0.542
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr15_-_54051837
|
0.537
|
NM_182758
|
WDR72
|
WD repeat domain 72
|
|
chrX_-_77914824
|
0.537
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr5_-_149792370
|
0.536
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr2_-_217540722
|
0.533
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr1_-_216978708
|
0.533
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_+_140165875
|
0.528
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr16_-_28634873
|
0.528
|
NM_177536
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_-_57234279
|
0.519
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr12_-_10151689
|
0.519
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr15_+_71184781
|
0.518
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr11_+_60467046
|
0.518
|
NM_031457
|
MS4A8B
|
membrane-spanning 4-domains, subfamily A, member 8B
|
|
chr14_+_21249209
|
0.517
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr4_+_15471488
|
0.517
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr1_-_27952591
|
0.515
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr18_-_45663667
|
0.514
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr16_+_28699878
|
0.510
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr7_+_37960920
|
0.506
|
NM_001242948
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr9_-_35563895
|
0.500
|
NM_001099951
NM_001164310
|
FAM166B
|
family with sequence similarity 166, member B
|
|
chr11_+_65268313
|
0.497
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr16_-_28437663
|
0.496
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr5_-_13944588
|
0.496
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr4_-_84035888
|
0.494
|
|
PLAC8
|
placenta-specific 8
|
|
chr12_-_21757752
|
0.491
|
NM_021957
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr19_+_3728373
|
0.490
|
NM_014428
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr7_-_55930444
|
0.486
|
NM_207366
|
SEPT14
|
septin 14
|
|
chr3_-_145968819
|
0.483
|
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr9_+_79115548
|
0.479
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr1_-_57045129
|
0.479
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_67001377
|
0.479
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr2_-_113012601
|
0.472
|
NM_032494
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr2_+_169757749
|
0.470
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr15_-_45670955
|
0.467
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr1_-_170043608
|
0.467
|
NM_001204514
NM_001204516
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr6_-_39072412
|
0.466
|
|
C6orf64
|
chromosome 6 open reading frame 64
|
|
chr11_-_108464344
|
0.466
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr3_-_145968958
|
0.465
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr7_+_90339146
|
0.464
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr18_+_28898051
|
0.462
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr18_-_24765178
|
0.461
|
NM_031422
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr4_-_84035908
|
0.460
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr7_+_90338711
|
0.460
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr4_+_108814419
|
0.458
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr11_-_26593667
|
0.457
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr1_+_104292439
|
0.450
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr11_+_124120422
|
0.449
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr8_-_110657745
|
0.448
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr22_+_24105207
|
0.440
|
NM_182520
|
C22orf15
|
chromosome 22 open reading frame 15
|
|
chr4_+_72204769
|
0.438
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_34195745
|
0.437
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr9_-_117267710
|
0.436
|
NM_001173425
NM_015404
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr1_-_162838550
|
0.436
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chr1_-_57044980
|
0.435
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr17_+_17876126
|
0.428
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr4_+_36285643
|
0.428
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr10_-_94003002
|
0.428
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr1_+_114496498
|
0.427
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr7_-_112726143
|
0.422
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr4_+_41614725
|
0.422
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_36549675
|
0.421
|
NM_014466
|
TEKT2
|
tektin 2 (testicular)
|
|
chr15_+_49462408
|
0.418
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr12_+_131438451
|
0.415
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr1_+_36748164
|
0.415
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr12_-_68726036
|
0.411
|
NM_001205028
NM_001205029
NM_017440
NM_020128
|
MDM1
|
Mdm1 nuclear protein homolog (mouse)
|
|
chr8_+_144731829
|
0.411
|
NM_014789
|
ZNF623
|
zinc finger protein 623
|
|
chr1_-_108786663
|
0.410
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chr14_-_34419957
|
0.410
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr11_-_107582733
|
0.408
|
NM_003063
|
SLN
|
sarcolipin
|