|
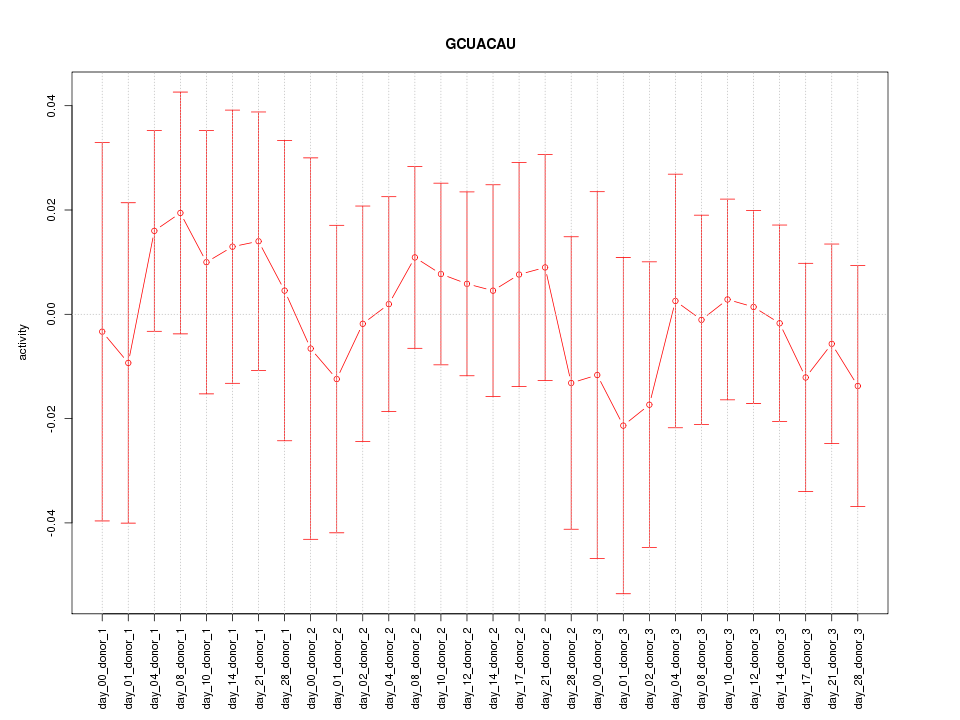
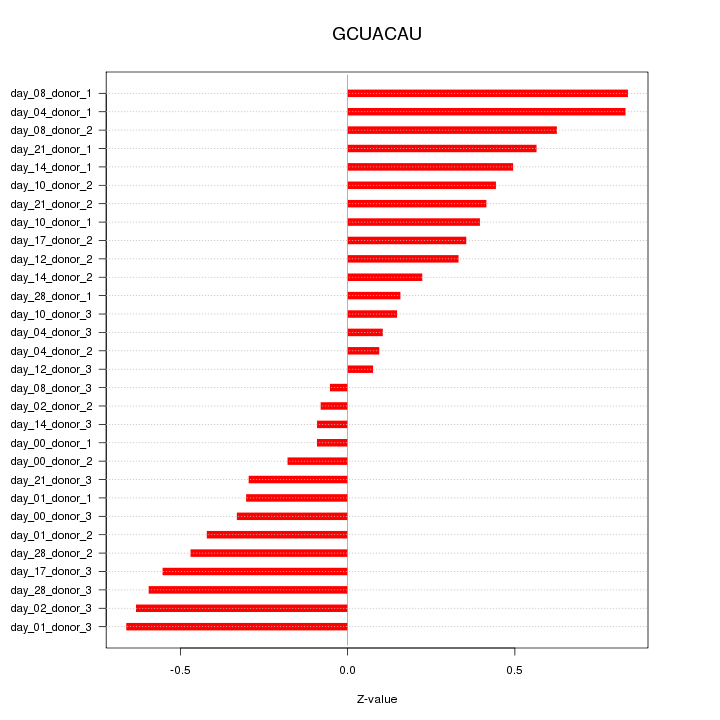
chr15_-_40398286
|
0.792
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr15_-_40398573
|
0.781
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr15_-_40401041
|
0.723
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr4_+_72052354
|
0.699
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr6_+_16129285
|
0.572
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr16_+_28303622
|
0.572
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr3_-_145968958
|
0.518
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr4_+_72204769
|
0.479
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_+_55523906
|
0.453
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr1_+_114496498
|
0.427
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr1_+_114493766
|
0.420
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chrX_-_44202834
|
0.375
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr17_-_47439326
|
0.357
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr7_+_114055048
|
0.338
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr6_-_56112315
|
0.331
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr5_+_140254930
|
0.307
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140213968
|
0.305
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr14_+_74111577
|
0.301
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr5_+_140220811
|
0.300
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_140186658
|
0.295
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr15_+_82555101
|
0.290
|
NM_001008226
|
FAM154B
|
family with sequence similarity 154, member B
|
|
chr16_+_15596122
|
0.288
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr10_+_120789227
|
0.284
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr13_-_36705513
|
0.279
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr3_+_3841079
|
0.262
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr13_-_36429798
|
0.260
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr5_+_140247767
|
0.253
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr2_-_213403280
|
0.253
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr2_-_24149899
|
0.247
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr1_-_204329043
|
0.247
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr16_+_15528324
|
0.237
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr7_+_94285620
|
0.232
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr1_-_156051788
|
0.230
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr7_+_94285676
|
0.228
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr16_+_69599868
|
0.221
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr18_-_53255734
|
0.220
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr12_+_12869818
|
0.219
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr2_-_170550859
|
0.215
|
NM_001085447
|
C2orf77
|
chromosome 2 open reading frame 77
|
|
chr8_+_28747885
|
0.212
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr8_-_52811707
|
0.207
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr5_+_140165875
|
0.207
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr10_+_93170086
|
0.205
|
NM_173497
NM_182765
|
HECTD2
|
HECT domain containing 2
|
|
chr15_+_57210823
|
0.200
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr7_+_121513158
|
0.198
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr6_+_17281773
|
0.193
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282931
|
0.190
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr5_+_140207562
|
0.188
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr6_+_17282294
|
0.188
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr8_-_110657745
|
0.186
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_-_110657025
|
0.183
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_-_110661007
|
0.182
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_-_152955502
|
0.180
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr3_+_183353355
|
0.178
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr8_-_110693233
|
0.178
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_-_110704019
|
0.177
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_56469655
|
0.175
|
NM_001203246
NM_022913
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr5_+_56509900
|
0.173
|
NM_001127236
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr2_-_152955208
|
0.173
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr8_-_110655418
|
0.172
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_111878450
|
0.172
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
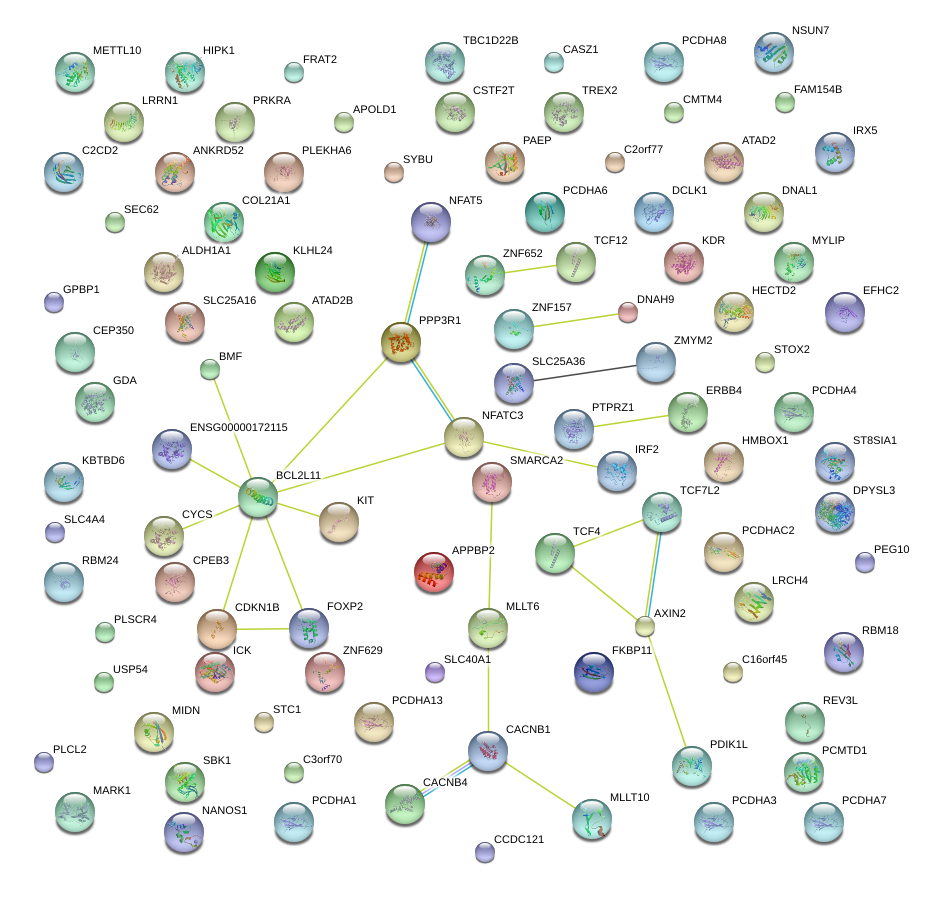
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr4_+_40751913
|
0.169
|
NM_024677
|
NSUN7
|
NOP2/Sun domain family, member 7
|
|
chr15_-_52970808
|
0.166
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr9_-_75568232
|
0.165
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_+_26438267
|
0.163
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr6_-_111804413
|
0.163
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr4_-_55991746
|
0.162
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr15_+_57511616
|
0.162
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr21_-_43373938
|
0.158
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr3_+_169684549
|
0.157
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr2_-_68479481
|
0.156
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr16_-_30798206
|
0.156
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr2_-_152830448
|
0.156
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_+_140345746
|
0.155
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr12_-_56652118
|
0.153
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr5_-_146889618
|
0.152
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr8_+_28748030
|
0.151
|
NM_024567
|
HMBOX1
|
homeobox containing 1
|
|
chr5_+_56471272
|
0.151
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr1_+_220701547
|
0.150
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr5_-_146833150
|
0.149
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr10_-_53459309
|
0.147
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr16_-_66730466
|
0.145
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr5_+_140180782
|
0.142
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr8_-_23712297
|
0.138
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr19_+_1248548
|
0.136
|
NM_177401
|
MIDN
|
midnolin
|
|
chr17_-_63557615
|
0.135
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr6_-_52926588
|
0.135
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr17_-_58603492
|
0.135
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr4_+_184826427
|
0.134
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr13_+_20532906
|
0.132
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr12_+_12938540
|
0.128
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr6_+_37225506
|
0.128
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr3_+_16926451
|
0.128
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr9_+_74729510
|
0.127
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chr7_-_25164902
|
0.127
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr10_-_94050799
|
0.127
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_-_27851744
|
0.124
|
NM_001142683
NM_024584
|
CCDC121
|
coiled-coil domain containing 121
|
|
chr5_+_140261792
|
0.123
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr13_-_41706587
|
0.123
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr16_+_68119211
|
0.122
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr13_+_20532788
|
0.118
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr2_-_190445521
|
0.115
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr17_+_36861857
|
0.115
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr9_+_2015218
|
0.113
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr3_+_140660639
|
0.110
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr1_+_179923870
|
0.110
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr16_+_54965110
|
0.109
|
NM_001252197
NM_005853
|
IRX5
|
iroquois homeobox 5
|
|
chr10_-_99094427
|
0.109
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr10_-_126480349
|
0.109
|
NM_212554
|
METTL10
|
methyltransferase like 10
|
|
chr12_-_22487566
|
0.108
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr2_-_179315756
|
0.107
|
NM_003690
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator
|
|
chr2_-_179315354
|
0.107
|
NM_001139518
NM_001139517
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator
|
|
chr1_-_10856704
|
0.105
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr3_-_184870727
|
0.104
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr4_-_185395616
|
0.104
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr9_-_125027080
|
0.103
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr1_-_89357246
|
0.100
|
NM_001514
|
GTF2B
|
general transcription factor IIB
|
|
chr9_-_15510899
|
0.099
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr21_-_43346780
|
0.099
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr15_-_56535475
|
0.098
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr2_-_20189775
|
0.098
|
NM_001006657
NM_020779
|
WDR35
|
WD repeat domain 35
|
|
chr10_-_94003002
|
0.096
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_+_16974581
|
0.095
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chr5_-_157285977
|
0.094
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr3_-_11761946
|
0.094
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr8_-_93978264
|
0.094
|
NM_001191035
NM_001171795
NM_001171796
NM_001171797
NM_001171798
NM_001171799
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr12_+_6898605
|
0.093
|
NM_000616
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr5_-_9546157
|
0.092
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr5_+_138677517
|
0.092
|
NM_016480
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr1_+_14075861
|
0.091
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr10_+_114709963
|
0.091
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr1_+_114471995
|
0.091
|
NM_152696
NM_198268
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr4_+_106816593
|
0.091
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr1_+_150980895
|
0.091
|
NM_021222
|
PRUNE
|
prune homolog (Drosophila)
|
|
chr18_+_11751470
|
0.090
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr8_+_79428335
|
0.090
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr11_-_74178593
|
0.090
|
NM_005472
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|
|
chr17_-_27621163
|
0.089
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr12_+_120427476
|
0.089
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr5_-_43557520
|
0.089
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr13_+_88324777
|
0.088
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr12_+_41831484
|
0.088
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr20_+_11898536
|
0.087
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr17_+_60556247
|
0.087
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr5_+_141488314
|
0.086
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr3_+_69812961
|
0.086
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_47476225
|
0.086
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_+_67584251
|
0.085
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr14_-_53019036
|
0.085
|
NM_001160047
NM_020784
|
TXNDC16
|
thioredoxin domain containing 16
|
|
chr14_-_36278335
|
0.085
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr5_+_67511578
|
0.085
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr20_-_40246991
|
0.084
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr12_+_41582167
|
0.083
|
NM_001164595
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr13_-_111567415
|
0.083
|
NM_017664
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chr18_-_19284737
|
0.083
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr5_+_79703792
|
0.082
|
NM_001105251
NM_014733
|
ZFYVE16
|
zinc finger, FYVE domain containing 16
|
|
chr18_-_25757351
|
0.082
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_-_160472960
|
0.082
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr9_-_37465384
|
0.082
|
NM_014872
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr11_+_45868956
|
0.082
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr9_-_112260529
|
0.081
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr1_+_74663895
|
0.081
|
NM_001112808
NM_001199327
NM_001199328
NM_001199329
NM_003838
|
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K readthrough
fucose-1-phosphate guanylyltransferase
|
|
chr11_+_45868668
|
0.080
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr16_+_19535178
|
0.080
|
NM_001199022
NM_014711
|
CCP110
|
centriolar coiled coil protein 110kDa
|
|
chr7_-_99679192
|
0.080
|
NM_017715
NM_032924
|
ZNF3
|
zinc finger protein 3
|
|
chr2_+_109335906
|
0.080
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr5_+_67586466
|
0.079
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_57045235
|
0.079
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chrX_+_17393537
|
0.079
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_86174088
|
0.078
|
NM_001170670
NM_017953
|
ZNHIT6
|
zinc finger, HIT-type containing 6
|
|
chr4_+_4291923
|
0.078
|
NM_145291
|
ZBTB49
|
zinc finger and BTB domain containing 49
|
|
chr2_+_149632772
|
0.078
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr9_+_96338671
|
0.077
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr5_+_140235594
|
0.076
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr1_-_155881163
|
0.076
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr3_-_11610264
|
0.076
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_-_20126368
|
0.075
|
NM_181719
|
TMCO4
|
transmembrane and coiled-coil domains 4
|
|
chrX_+_17653412
|
0.075
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_224033673
|
0.075
|
NM_001031685
NM_005426
|
TP53BP2
|
tumor protein p53 binding protein, 2
|
|
chr3_-_11685395
|
0.074
|
NM_001128219
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr8_-_27472298
|
0.074
|
NM_001831
|
CLU
|
clusterin
|
|
chr2_-_100106478
|
0.073
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr11_+_100557851
|
0.073
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr3_+_69915374
|
0.073
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr7_+_23636970
|
0.073
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr21_+_40177847
|
0.073
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr17_+_38171613
|
0.072
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr3_-_11623829
|
0.071
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_46806389
|
0.071
|
NM_199044
|
NSUN4
|
NOP2/Sun domain family, member 4
|
|
chr5_+_67588395
|
0.071
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_161694992
|
0.071
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr4_-_89618928
|
0.071
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr10_-_75335415
|
0.071
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr6_+_90539569
|
0.069
|
NM_001137667
NM_001137668
NM_012115
|
CASP8AP2
|
caspase 8 associated protein 2
|
|
chr3_+_69812620
|
0.068
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_74383301
|
0.068
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr20_+_56964162
|
0.067
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr5_-_175964223
|
0.067
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr13_-_86373328
|
0.067
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr11_+_48002063
|
0.067
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr3_+_69788562
|
0.067
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|