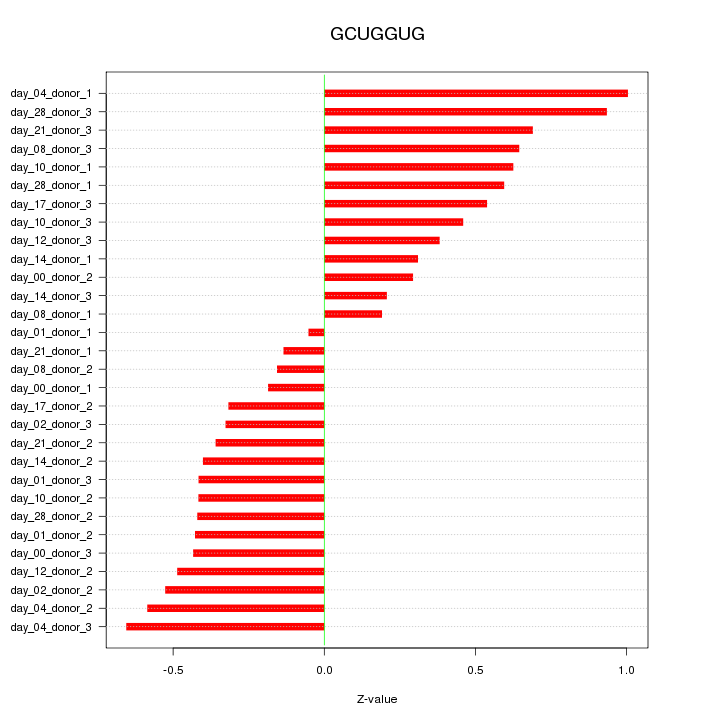
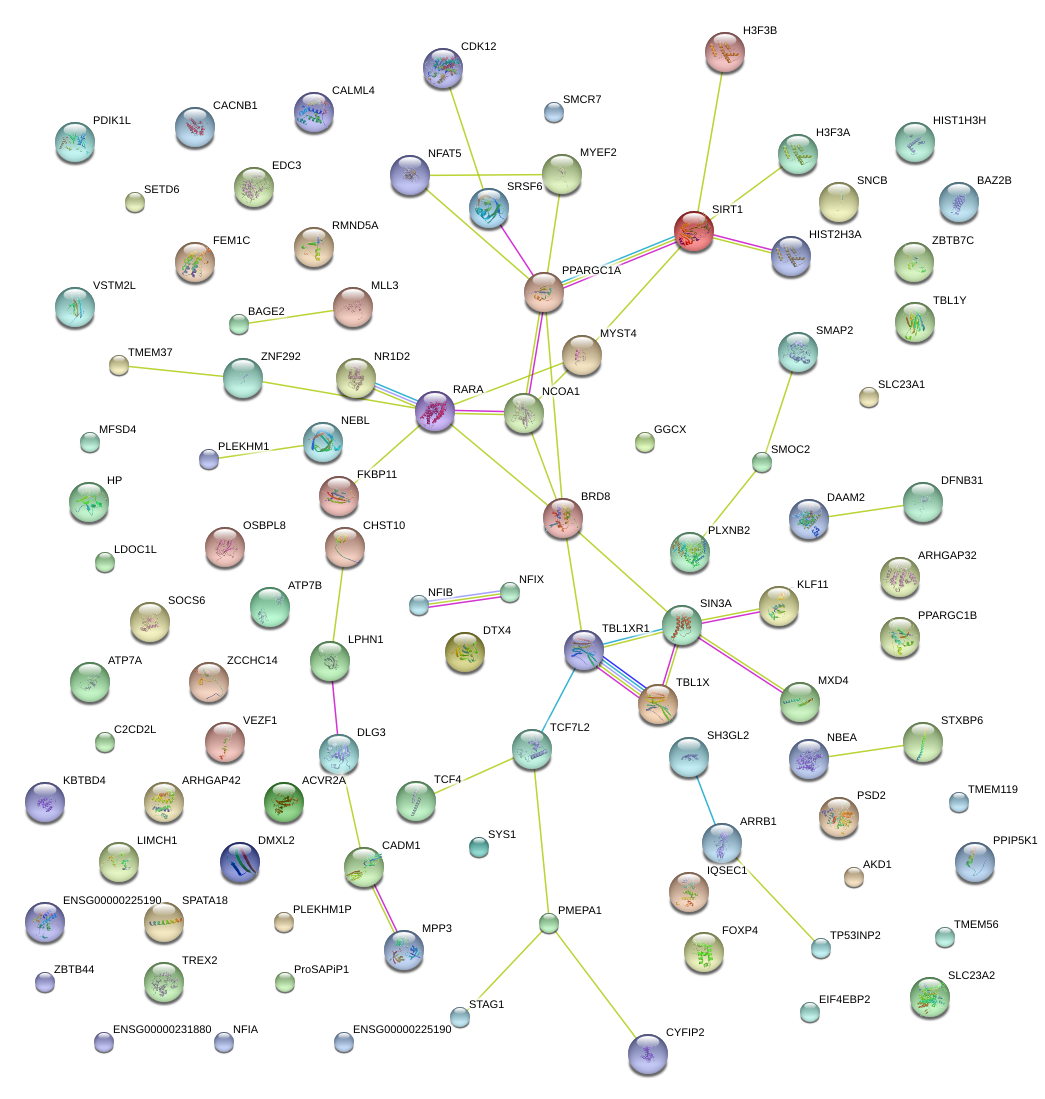
|
chr13_+_36050885
|
0.753
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr15_-_68498373
|
0.741
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr5_+_156696351
|
0.706
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr5_+_156693051
|
0.667
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr13_+_35516390
|
0.631
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr9_+_17578952
|
0.606
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr17_+_38498270
|
0.563
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr20_+_36531498
|
0.547
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr20_-_3149070
|
0.489
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_+_40859016
|
0.484
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr1_+_40839368
|
0.472
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr6_+_39760148
|
0.470
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr6_+_39760772
|
0.453
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr2_+_10184371
|
0.451
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr1_+_40840304
|
0.446
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr1_+_61547625
|
0.444
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61542928
|
0.416
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_40862471
|
0.415
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr4_+_52917440
|
0.406
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr4_+_41614911
|
0.403
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_61547388
|
0.382
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_10183630
|
0.378
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr4_+_41362750
|
0.373
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr20_+_43990576
|
0.352
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr19_+_13105970
|
0.350
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_108991893
|
0.346
|
NM_181724
|
TMEM119
|
transmembrane protein 119
|
|
chr3_-_13008959
|
0.325
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr15_-_43877043
|
0.322
|
NM_014659
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr19_-_14316980
|
0.314
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr13_-_52585472
|
0.306
|
NM_000053
NM_001005918
NM_001243182
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr5_-_114880536
|
0.304
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr11_+_58939539
|
0.296
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr4_-_23891608
|
0.291
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr20_+_42086468
|
0.290
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr11_+_100557851
|
0.284
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr15_-_43882387
|
0.283
|
NM_001130858
NM_001130859
NM_001190214
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr18_+_67956136
|
0.278
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr5_-_176057324
|
0.270
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr20_+_54933982
|
0.268
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chrX_+_69674914
|
0.268
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chrX_+_9431314
|
0.263
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr18_-_45663667
|
0.255
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr15_-_48470453
|
0.255
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr1_+_26438267
|
0.255
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr11_-_115375065
|
0.255
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr18_-_53255734
|
0.253
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chrX_+_69664704
|
0.251
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr10_+_76586299
|
0.249
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr2_+_86947387
|
0.247
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr11_+_118978087
|
0.246
|
NM_014807
|
C2CD2L
|
C2CD2-like
|
|
chr16_+_58549382
|
0.246
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr2_+_148602569
|
0.242
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr3_-_136471186
|
0.239
|
NM_005862
|
STAG1
|
stromal antigen 1
|
|
chr7_-_152133071
|
0.239
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr2_+_120189342
|
0.237
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr10_+_72163860
|
0.236
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr15_-_51914770
|
0.235
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr10_+_69644938
|
0.233
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr11_-_47600458
|
0.230
|
NM_016506
NM_018095
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4
|
|
chr15_-_74988342
|
0.227
|
NM_001142443
NM_001142444
NM_025083
|
EDC3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae)
|
|
chr2_-_85788644
|
0.225
|
NM_000821
NM_001142269
|
GGCX
|
gamma-glutamyl carboxylase
|
|
chr16_+_69599868
|
0.222
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr17_-_56065596
|
0.220
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr6_+_41514133
|
0.220
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr6_+_87865261
|
0.219
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chrX_+_69672110
|
0.216
|
NM_020730
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr11_-_130184459
|
0.212
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr2_-_160472960
|
0.212
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr5_+_149151502
|
0.210
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr11_-_129062092
|
0.210
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr3_+_23987514
|
0.208
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr5_+_139175385
|
0.207
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr2_+_24807345
|
0.206
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr1_+_95558072
|
0.201
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr1_+_205538111
|
0.201
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr22_-_44893903
|
0.201
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr15_-_75743776
|
0.200
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr5_+_149109814
|
0.198
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr2_-_101034049
|
0.197
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr3_-_13114547
|
0.196
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr16_-_87525317
|
0.195
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr15_-_75744029
|
0.194
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr10_-_21186530
|
0.183
|
NM_006393
|
NEBL
|
nebulette
|
|
chr11_-_75062650
|
0.183
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr17_-_73775822
|
0.182
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr3_+_23986735
|
0.179
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr15_-_75748123
|
0.178
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr10_+_69644341
|
0.177
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr9_-_117267710
|
0.173
|
NM_001173425
NM_015404
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr14_-_25519076
|
0.172
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr17_+_37617688
|
0.170
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr20_+_33292117
|
0.170
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr17_+_18164423
|
0.170
|
NM_148886
|
SMCR7
|
Smith-Magenis syndrome chromosome region, candidate 7
|
|
chrX_+_9432980
|
0.170
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_-_4982133
|
0.168
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr17_+_18163836
|
0.167
|
NM_001144900
NM_139162
|
SMCR7
|
Smith-Magenis syndrome chromosome region, candidate 7
|
|
chr4_-_2263706
|
0.167
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr17_-_43568124
|
0.166
|
NM_014798
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr22_-_50746000
|
0.165
|
NM_012401
|
PLXNB2
|
plexin B2
|
|
chr9_+_115513050
|
0.164
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr11_+_66234293
|
0.163
|
NM_001098510
NM_001243136
NM_145065
|
PELI3
|
pellino homolog 3 (Drosophila)
|
|
chr9_-_117265452
|
0.162
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr10_-_12085022
|
0.162
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr10_-_12084772
|
0.161
|
NM_015542
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr10_-_21463019
|
0.159
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr2_-_97535669
|
0.157
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr4_-_185570575
|
0.156
|
NM_004346
NM_032991
|
CASP3
|
caspase 3, apoptosis-related cysteine peptidase
|
|
chrX_-_53254603
|
0.155
|
NM_001146702
NM_004187
|
KDM5C
|
lysine (K)-specific demethylase 5C
|
|
chr17_+_74868728
|
0.155
|
NM_198955
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr1_+_114496498
|
0.155
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr6_-_100911550
|
0.153
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr17_-_58603492
|
0.152
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr15_+_57511616
|
0.151
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr15_+_57210823
|
0.151
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr2_+_85581840
|
0.150
|
NM_001135021
NM_001135022
NM_001135023
NM_032213
|
ELMOD3
|
ELMO/CED-12 domain containing 3
|
|
chr10_+_18948276
|
0.149
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr1_+_114493766
|
0.148
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr9_-_111882209
|
0.148
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr11_+_11862955
|
0.147
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr4_+_159131400
|
0.146
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr3_-_142166782
|
0.143
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr9_-_131534186
|
0.142
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr18_+_46065367
|
0.141
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr19_+_47421932
|
0.140
|
NM_004491
|
ARHGAP35
|
Rho GTPase activating protein 35
|
|
chr17_+_7743233
|
0.139
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr12_-_30830143
|
0.138
|
NM_001190995
|
IPO8
|
importin 8
|
|
chr9_-_36401149
|
0.137
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr7_-_131241321
|
0.137
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr2_+_14772809
|
0.135
|
NM_145175
|
FAM84A
|
family with sequence similarity 84, member A
|
|
chr3_-_33481877
|
0.134
|
NM_001128161
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr17_+_36861857
|
0.134
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr17_-_3749455
|
0.133
|
NM_001114118
NM_018553
|
C17orf85
|
chromosome 17 open reading frame 85
|
|
chr8_-_52811707
|
0.133
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr11_-_70507911
|
0.132
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr19_-_11670003
|
0.132
|
NM_032377
|
ELOF1
|
elongation factor 1 homolog (S. cerevisiae)
|
|
chr14_-_75179739
|
0.130
|
NM_001039479
|
KIAA0317
|
KIAA0317
|
|
chr1_-_41131323
|
0.130
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr15_+_49170264
|
0.129
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr3_-_49823626
|
0.128
|
NM_001242829
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr1_+_95582827
|
0.127
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr15_-_75199461
|
0.127
|
NM_020447
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr1_+_162351519
|
0.127
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr6_+_4021500
|
0.126
|
NM_003913
|
PRPF4B
|
PRP4 pre-mRNA processing factor 4 homolog B (yeast)
|
|
chr4_-_78740508
|
0.125
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr3_-_33481862
|
0.124
|
NM_001128160
NM_014517
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr2_+_217498082
|
0.122
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr16_+_71560022
|
0.121
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr8_-_116681103
|
0.120
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr5_+_140044383
|
0.120
|
NM_017706
|
WDR55
|
WD repeat domain 55
|
|
chr7_+_8008416
|
0.119
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr18_-_57026465
|
0.118
|
NM_005570
|
LMAN1
|
lectin, mannose-binding, 1
|
|
chr9_-_36400212
|
0.118
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr6_-_31628509
|
0.118
|
NM_021184
|
C6orf47
|
chromosome 6 open reading frame 47
|
|
chr10_+_129845806
|
0.117
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr22_+_41697452
|
0.117
|
NM_017590
|
ZC3H7B
|
zinc finger CCCH-type containing 7B
|
|
chr11_-_67980699
|
0.117
|
NM_016028
NM_017635
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila)
|
|
chr20_+_43991739
|
0.116
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 readthrough
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr10_+_181410
|
0.116
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr19_+_45909466
|
0.115
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr20_+_47538211
|
0.114
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr17_-_6459746
|
0.114
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr20_+_32150139
|
0.111
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr17_+_74864427
|
0.111
|
NM_001199172
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr9_-_130679210
|
0.110
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr13_+_77566043
|
0.109
|
NM_006493
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5
|
|
chr10_-_65028825
|
0.108
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chrX_-_102531673
|
0.108
|
NM_001012979
|
TCEAL5
|
transcription elongation factor A (SII)-like 5
|
|
chr9_-_36400817
|
0.108
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr7_+_90338711
|
0.107
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr3_+_51705221
|
0.106
|
NM_001129884
NM_001243727
NM_015926
NM_001243725
NM_001243726
|
TEX264
|
testis expressed 264
|
|
chr1_+_210406194
|
0.106
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr3_+_42544094
|
0.105
|
NM_001251883
NM_001251884
NM_001251885
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr1_+_36273786
|
0.105
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr17_+_72983723
|
0.104
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr16_+_50186809
|
0.104
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr14_+_60712469
|
0.104
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr5_-_56247915
|
0.102
|
NM_152622
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr2_+_28615771
|
0.101
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr10_-_73533196
|
0.101
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_-_109935869
|
0.100
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr9_-_33264458
|
0.100
|
NM_001172415
NM_004323
|
BAG1
|
BCL2-associated athanogene
|
|
chr20_-_3996035
|
0.100
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr20_-_4990913
|
0.100
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chrX_-_25034064
|
0.099
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr17_+_4402128
|
0.097
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr15_+_44719578
|
0.097
|
NM_016396
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr7_+_129007942
|
0.097
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chrX_+_152599544
|
0.096
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr5_+_167718485
|
0.096
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr7_+_128864740
|
0.095
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_128894008
|
0.095
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr15_+_64791618
|
0.094
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chr5_-_90679115
|
0.093
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr17_-_9940004
|
0.093
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr19_+_51815095
|
0.092
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr19_+_7968764
|
0.092
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr16_-_57318574
|
0.091
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr3_+_49449638
|
0.091
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr3_-_49823972
|
0.091
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr20_+_816710
|
0.091
|
NM_207121
|
FAM110A
|
family with sequence similarity 110, member A
|