|
chr6_-_35696359
|
0.609
|
NM_001145775
|
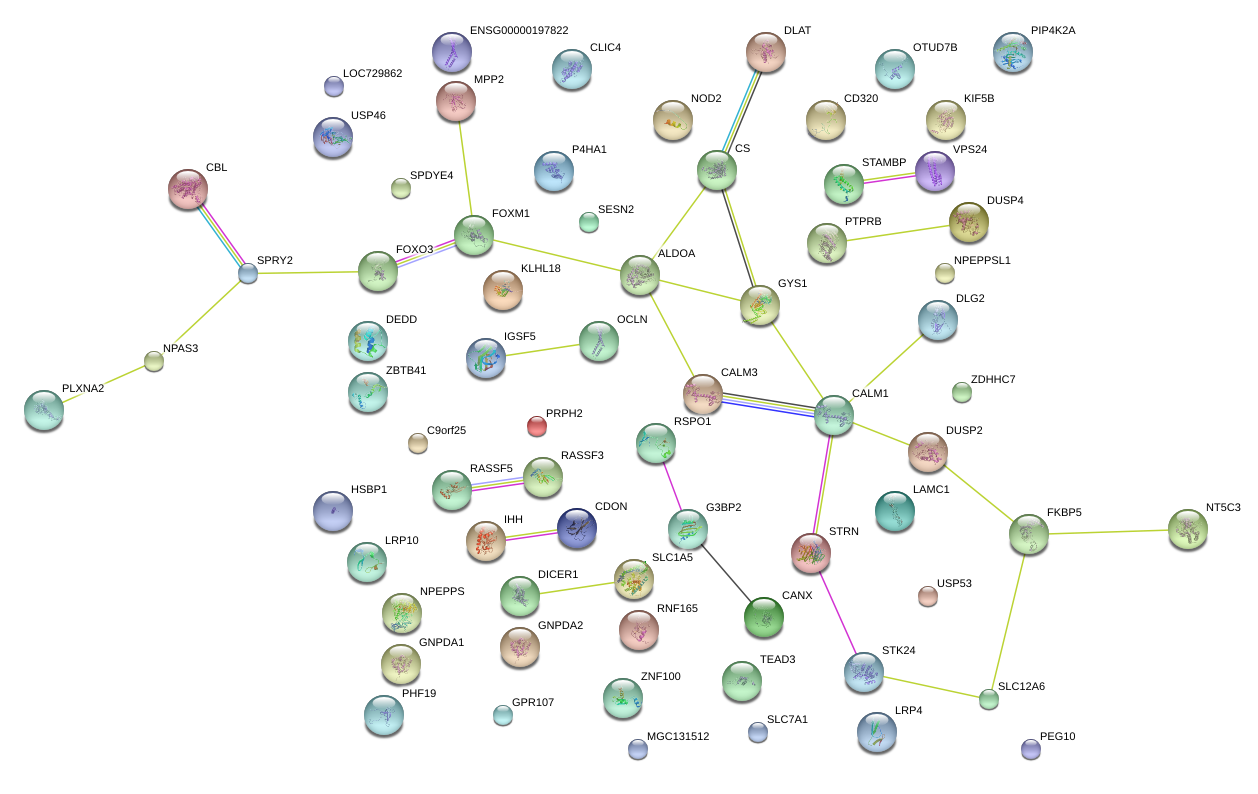
FKBP5
|
FK506 binding protein 5
|
|
chr6_-_35656677
|
0.440
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr10_-_74856656
|
0.379
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|
|
chr1_-_208417635
|
0.343
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr15_-_34630203
|
0.327
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_34629941
|
0.324
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_34629044
|
0.292
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr8_-_29206321
|
0.285
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr15_-_34610928
|
0.284
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr11_+_119076974
|
0.269
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr16_+_30064410
|
0.265
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr19_+_47104490
|
0.263
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr9_-_34458477
|
0.246
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr10_-_23003443
|
0.235
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr1_+_25071759
|
0.226
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr8_-_29207795
|
0.218
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr19_-_49496566
|
0.211
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr3_+_47324329
|
0.200
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr17_+_45608443
|
0.193
|
NM_006310
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr1_+_28585959
|
0.189
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr9_-_123639566
|
0.185
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr13_-_80915041
|
0.174
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr16_+_30075530
|
0.167
|
NM_184043
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr5_-_141392537
|
0.150
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr16_+_30075715
|
0.148
|
NM_001127617
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr19_-_8373157
|
0.142
|
NM_001165895
NM_016579
|
CD320
|
CD320 molecule
|
|
chr11_-_83984230
|
0.133
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr11_-_85338313
|
0.131
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr11_-_83393436
|
0.130
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr11_-_84028379
|
0.126
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_-_47290541
|
0.121
|
NM_001145144
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr9_+_132815984
|
0.112
|
NM_001136557
NM_001136558
NM_020960
|
GPR107
|
G protein-coupled receptor 107
|
|
chr1_-_38100575
|
0.112
|
NM_001038633
NM_001242908
NM_001242909
NM_001242910
|
RSPO1
|
R-spondin 1
|
|
chr19_-_47291594
|
0.109
|
NM_005628
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr1_+_182992329
|
0.103
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr4_+_120133764
|
0.103
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr11_-_84634437
|
0.101
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_+_94285620
|
0.091
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr19_-_47287998
|
0.091
|
NM_001145145
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr7_+_94285676
|
0.086
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr2_-_219925237
|
0.075
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr12_-_71031174
|
0.068
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr2_-_96811138
|
0.067
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr14_-_95599794
|
0.064
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_-_37193609
|
0.060
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr11_+_111895537
|
0.059
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr1_-_161102210
|
0.054
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr5_+_68800003
|
0.054
|
NM_001205255
|
OCLN
|
occludin
|
|
chr1_-_161102457
|
0.053
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr14_-_95608054
|
0.050
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr14_+_33408458
|
0.049
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr21_+_41117333
|
0.043
|
NM_001080444
|
IGSF5
|
immunoglobulin superfamily, member 5
|
|
chr13_-_99174251
|
0.042
|
NM_003576
|
STK24
|
serine/threonine kinase 24
|
|
chr16_+_50731049
|
0.041
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr12_-_71003622
|
0.040
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr18_+_43914186
|
0.040
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr13_-_99229226
|
0.039
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr4_-_76598289
|
0.037
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr4_-_76598611
|
0.037
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr2_+_74056042
|
0.037
|
NM_201647
|
STAMBP
|
STAM binding protein
|
|
chr2_+_74056095
|
0.036
|
NM_006463
NM_213622
|
STAMBP
|
STAM binding protein
|
|
chr4_-_44728518
|
0.031
|
NM_138335
|
GNPDA2
|
glucosamine-6-phosphate deaminase 2
|
|
chr16_-_85045119
|
0.027
|
NM_001145548
NM_017740
|
ZDHHC7
|
zinc finger, DHHC-type containing 7
|
|
chr5_+_68788118
|
0.026
|
NM_002538
|
OCLN
|
occludin
|
|
chr5_+_179125796
|
0.026
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr1_+_206680863
|
0.026
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr13_-_30169795
|
0.024
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr12_-_56694174
|
0.023
|
NM_004077
|
CS
|
citrate synthase
|
|
chr5_+_68788387
|
0.023
|
NM_001205254
|
OCLN
|
occludin
|
|
chr7_-_33102174
|
0.022
|
NM_001002010
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr16_+_30076993
|
0.018
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr2_-_86948244
|
0.017
|
NM_001198954
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough
|
|
chr14_+_23340955
|
0.014
|
NM_014045
|
LRP10
|
low density lipoprotein receptor-related protein 10
|
|
chr1_+_206730456
|
0.012
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr1_-_149982528
|
0.010
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr6_+_108881010
|
0.010
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr7_-_33080517
|
0.009
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr10_-_32345337
|
0.008
|
NM_004521
|
KIF5B
|
kinesin family member 5B
|
|
chr2_-_86790533
|
0.007
|
NM_001005753
NM_001193517
NM_016079
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr1_-_197169635
|
0.007
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr6_-_35464716
|
0.006
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr6_+_108882068
|
0.001
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr4_-_53522756
|
0.001
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|