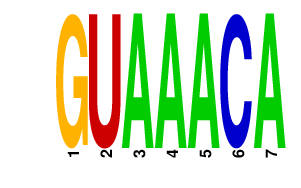
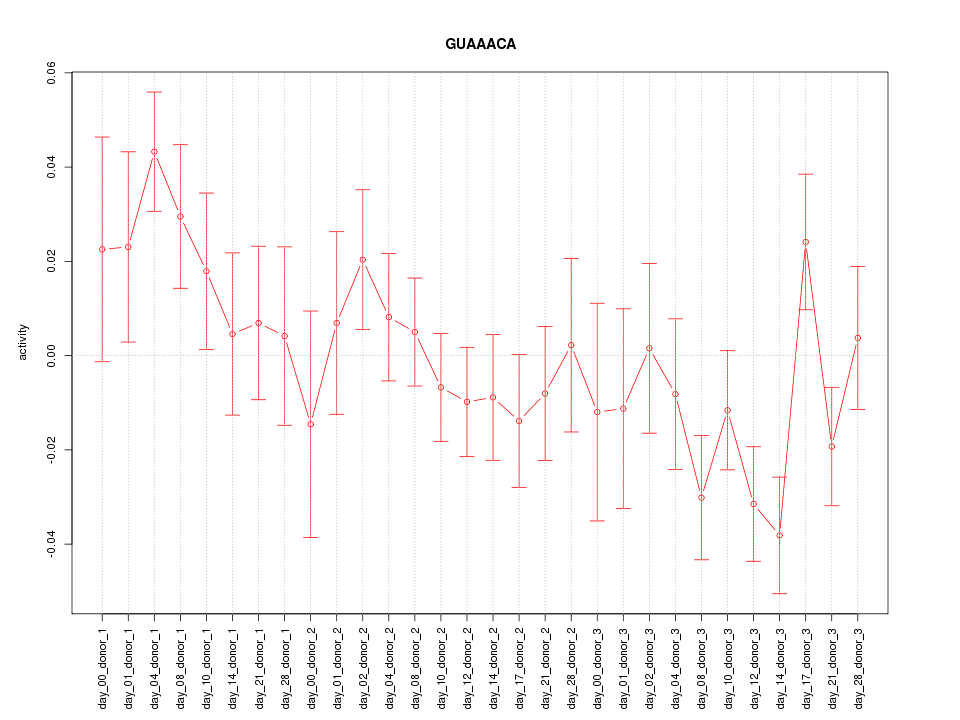
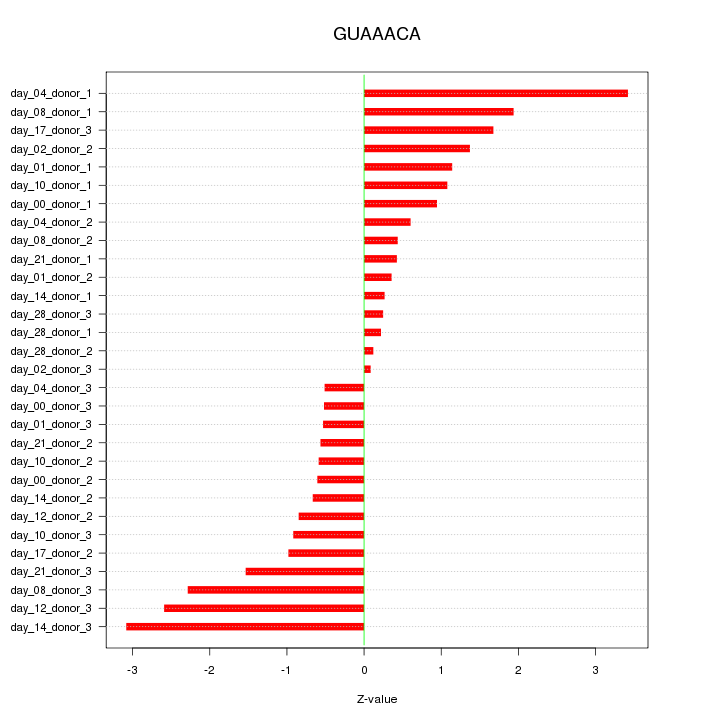
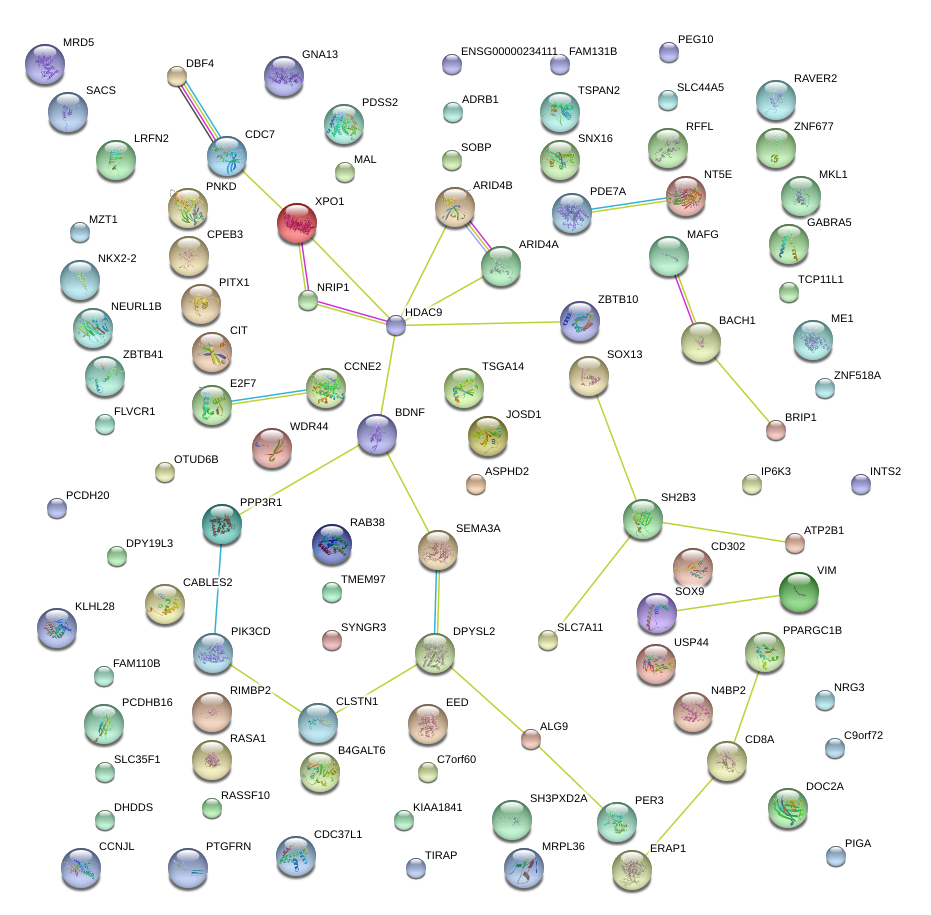
|
chr20_-_21494663
|
1.904
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr4_-_139163222
|
1.754
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr8_+_26371461
|
1.597
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr6_+_86159301
|
1.318
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr8_+_26435339
|
1.299
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_+_92082423
|
1.127
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chr5_+_172068188
|
1.093
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr12_-_77459198
|
1.074
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr7_+_18126543
|
1.020
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_18548899
|
0.999
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr1_+_91966365
|
0.946
|
NM_001134419
NM_003503
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr1_+_91966653
|
0.942
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr13_-_61989654
|
0.940
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr9_-_27573444
|
0.933
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr16_+_2039945
|
0.915
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr7_+_87505543
|
0.905
|
NM_006716
|
DBF4
|
DBF4 homolog (S. cerevisiae)
|
|
chr7_+_94285620
|
0.893
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr7_+_94285676
|
0.856
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr5_-_159739482
|
0.815
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr10_+_17270257
|
0.800
|
NM_003380
|
VIM
|
vimentin
|
|
chr13_-_73301726
|
0.767
|
NM_001071775
|
MZT1
|
mitotic spindle organizing protein 1
|
|
chr11_+_13030879
|
0.753
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr1_-_115632047
|
0.740
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr14_-_45431121
|
0.728
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr8_-_95907423
|
0.721
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr1_+_7844713
|
0.710
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr12_+_111843743
|
0.707
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr11_-_27741192
|
0.702
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721975
|
0.698
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr17_+_26646120
|
0.694
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr8_+_58906968
|
0.678
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr4_+_175204827
|
0.677
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr8_+_81398416
|
0.675
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr5_+_149151502
|
0.672
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr16_-_30022300
|
0.667
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr11_-_27742224
|
0.664
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr18_-_29264394
|
0.662
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr11_-_87908551
|
0.658
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr11_-_27721179
|
0.648
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27743604
|
0.646
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_65210724
|
0.646
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr5_+_149109814
|
0.638
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr11_-_27722599
|
0.634
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_-_66701175
|
0.630
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr10_+_97889467
|
0.629
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr11_-_27681195
|
0.628
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_86564727
|
0.627
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr11_-_27722446
|
0.622
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_90049818
|
0.620
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr11_-_27723061
|
0.615
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_-_84140854
|
0.612
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr6_+_107811272
|
0.609
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr6_+_118228688
|
0.606
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr7_-_112579726
|
0.604
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr8_-_82754427
|
0.602
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr17_-_63052884
|
0.595
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr11_-_111741980
|
0.577
|
NM_001077691
NM_001077692
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr5_+_86564044
|
0.575
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr12_-_120315074
|
0.574
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr7_-_106301329
|
0.573
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr7_-_143059696
|
0.568
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chrX_-_15353675
|
0.566
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr1_+_213031596
|
0.565
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr2_+_61292999
|
0.563
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr22_+_26825220
|
0.554
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr1_-_197169635
|
0.552
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr8_-_66753968
|
0.547
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr20_-_60982334
|
0.546
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr15_+_27112106
|
0.536
|
NM_001165037
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr1_-_76076762
|
0.535
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr2_+_95691428
|
0.519
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr4_+_40058507
|
0.513
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr7_-_83824216
|
0.513
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr7_-_143059144
|
0.511
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr10_+_115803805
|
0.510
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr10_-_105614952
|
0.508
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr21_-_16437125
|
0.507
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr7_-_130080823
|
0.504
|
NM_018718
|
CEP41
|
centrosomal protein 41kDa
|
|
chr6_-_40555111
|
0.500
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr1_+_204042191
|
0.498
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr2_-_68479481
|
0.497
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr15_+_27111865
|
0.496
|
NM_000810
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr12_-_131002409
|
0.495
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr1_+_9711781
|
0.494
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr6_-_107780682
|
0.493
|
NM_020381
|
PDSS2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2
|
|
chrX_+_117480027
|
0.486
|
NM_001184965
NM_001184966
NM_019045
|
WDR44
|
WD repeat domain 44
|
|
chr9_+_4679557
|
0.479
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr17_-_33390676
|
0.478
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr5_-_134369841
|
0.478
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_+_70117154
|
0.471
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr10_-_94050799
|
0.470
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr21_+_30671217
|
0.466
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr13_-_24007822
|
0.461
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr2_-_61765394
|
0.448
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr22_-_39095991
|
0.446
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr14_+_58765209
|
0.446
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr10_-_94003002
|
0.445
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr19_+_32897021
|
0.444
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr2_-_160654736
|
0.444
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr21_+_30671736
|
0.443
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr7_+_18535351
|
0.443
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr17_-_79881304
|
0.442
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr11_+_85955789
|
0.437
|
NM_003797
NM_152991
|
EED
|
embryonic ectoderm development
|
|
chr1_+_117452359
|
0.434
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr12_-_95945226
|
0.430
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr17_-_60005309
|
0.430
|
NM_020748
|
INTS2
|
integrator complex subunit 2
|
|
chr6_-_33714681
|
0.429
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr19_+_32896515
|
0.425
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr5_-_96143689
|
0.425
|
NM_001040458
NM_016442
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr11_+_33060962
|
0.423
|
NM_018393
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr10_+_83637442
|
0.417
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr2_+_219187858
|
0.416
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr16_-_3285359
|
0.414
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr12_-_95942570
|
0.413
|
NM_001042403
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr13_-_113242467
|
0.410
|
NM_006322
|
TUBGCP3
|
tubulin, gamma complex associated protein 3
|
|
chr11_+_33061560
|
0.409
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr12_-_131323616
|
0.406
|
NM_001980
NM_194356
|
STX2
|
syntaxin 2
|
|
chr9_-_72287097
|
0.406
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr9_-_14314036
|
0.402
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_206138909
|
0.402
|
NM_001123168
|
FAM72B
FAM72A
|
family with sequence similarity 72, member B
family with sequence similarity 72, member A
|
|
chr20_+_42295708
|
0.401
|
NM_002466
|
MYBL2
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 2
|
|
chrX_+_46771599
|
0.392
|
NM_001077445
NM_014735
|
PHF16
|
PHD finger protein 16
|
|
chr2_-_26205437
|
0.391
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr5_-_132073143
|
0.391
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr22_-_37823503
|
0.387
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr12_+_104359557
|
0.386
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr10_+_83635069
|
0.386
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr3_+_180630054
|
0.384
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr7_-_2354050
|
0.382
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr2_-_201828180
|
0.381
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr6_+_72926462
|
0.381
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_182970853
|
0.377
|
NM_032047
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr9_-_14398981
|
0.377
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr16_+_66914380
|
0.376
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr2_+_32288579
|
0.374
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr1_-_24513737
|
0.373
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr4_+_37585854
|
0.371
|
NM_018302
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr2_-_166650770
|
0.371
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr19_+_34745452
|
0.371
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr6_+_150464156
|
0.368
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr3_+_142375738
|
0.364
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr11_-_111742205
|
0.363
|
NM_001077690
NM_024740
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr1_+_114493766
|
0.362
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr18_-_43652153
|
0.362
|
NM_024430
|
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2
|
|
chr1_+_114496498
|
0.361
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr2_-_24149899
|
0.361
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr4_-_37687932
|
0.360
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr16_+_75032874
|
0.357
|
NM_032268
|
ZNRF1
|
zinc and ring finger 1
|
|
chr6_-_42419782
|
0.357
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr5_+_52284903
|
0.356
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr6_-_53213747
|
0.353
|
NM_001242828
NM_001242830
NM_001242831
NM_021814
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr17_+_42634598
|
0.353
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr19_-_344778
|
0.352
|
NM_017550
|
MIER2
|
mesoderm induction early response 1, family member 2
|
|
chrX_+_17393537
|
0.349
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr3_+_58318597
|
0.348
|
NM_017771
|
PXK
|
PX domain containing serine/threonine kinase
|
|
chr5_+_49962771
|
0.348
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr7_+_35840595
|
0.348
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr9_+_103235489
|
0.346
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr1_-_109506016
|
0.346
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr8_+_67624652
|
0.345
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr4_+_37455551
|
0.338
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr14_+_103851693
|
0.338
|
NM_001128918
NM_001128919
NM_001128920
NM_001128921
NM_002376
|
MARK3
|
MAP/microtubule affinity-regulating kinase 3
|
|
chr10_+_69644938
|
0.336
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr4_-_52904424
|
0.336
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr10_-_43762366
|
0.335
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr20_+_48429249
|
0.334
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr1_-_207224298
|
0.334
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr12_+_57916607
|
0.332
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr6_-_83903632
|
0.331
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr15_-_34630203
|
0.329
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr8_+_87354958
|
0.328
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr14_-_69446038
|
0.327
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr6_-_119031230
|
0.327
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr1_+_90098643
|
0.327
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr7_-_23053732
|
0.325
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr8_+_67687415
|
0.325
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr6_+_106534188
|
0.323
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr10_+_69644341
|
0.322
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr14_-_61115906
|
0.322
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr2_+_153574385
|
0.321
|
NM_152522
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6
|
|
chr2_+_102314162
|
0.319
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr15_-_83953465
|
0.319
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr12_+_104458235
|
0.318
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr1_+_26737232
|
0.318
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr8_+_15397595
|
0.317
|
NM_006765
NM_178234
|
TUSC3
|
tumor suppressor candidate 3
|
|
chr6_+_45389818
|
0.316
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_+_17653412
|
0.315
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr7_-_50800018
|
0.315
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_-_197036301
|
0.314
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chrX_-_131547536
|
0.313
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_-_79787939
|
0.312
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr1_+_120839004
|
0.311
|
NM_001100910
|
FAM72B
|
family with sequence similarity 72, member B
|
|
chr3_-_27498244
|
0.310
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr22_+_30792929
|
0.309
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr7_-_111846384
|
0.309
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr7_+_100770378
|
0.306
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr2_-_219432955
|
0.306
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr15_-_34629044
|
0.306
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_199996729
|
0.305
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr2_+_48541767
|
0.305
|
NM_002158
|
FOXN2
|
forkhead box N2
|