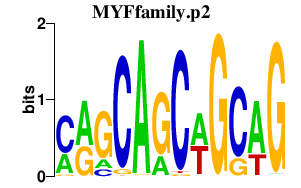
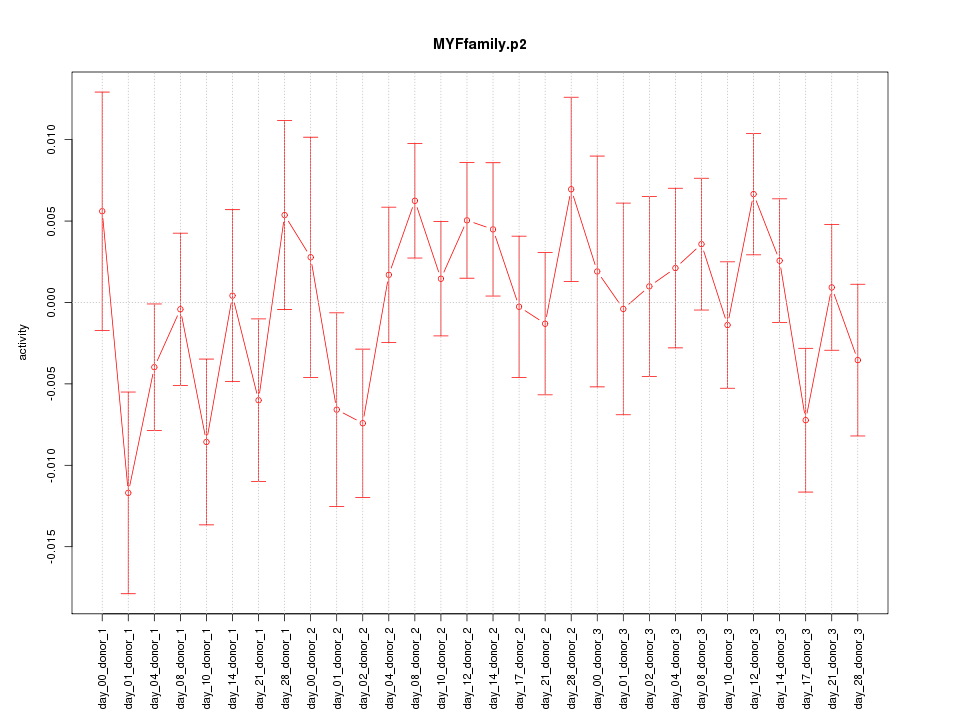
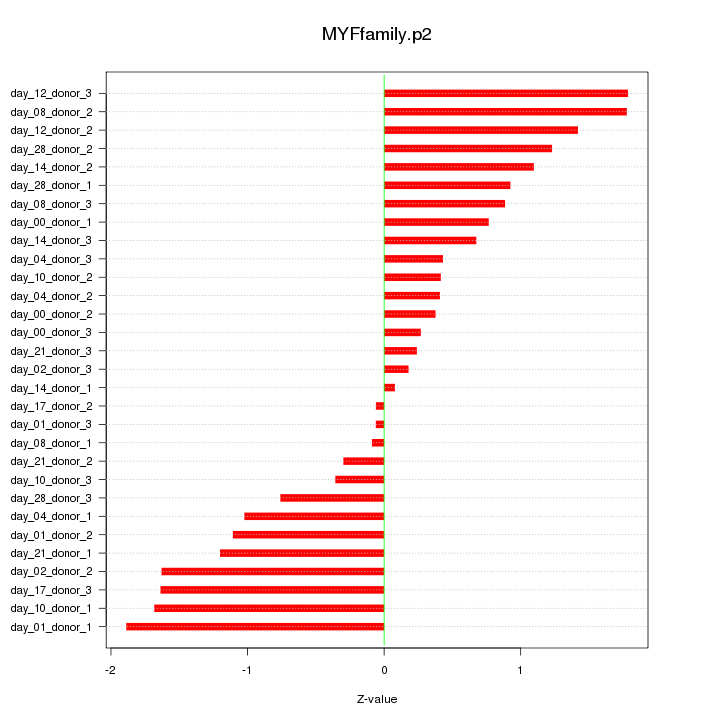
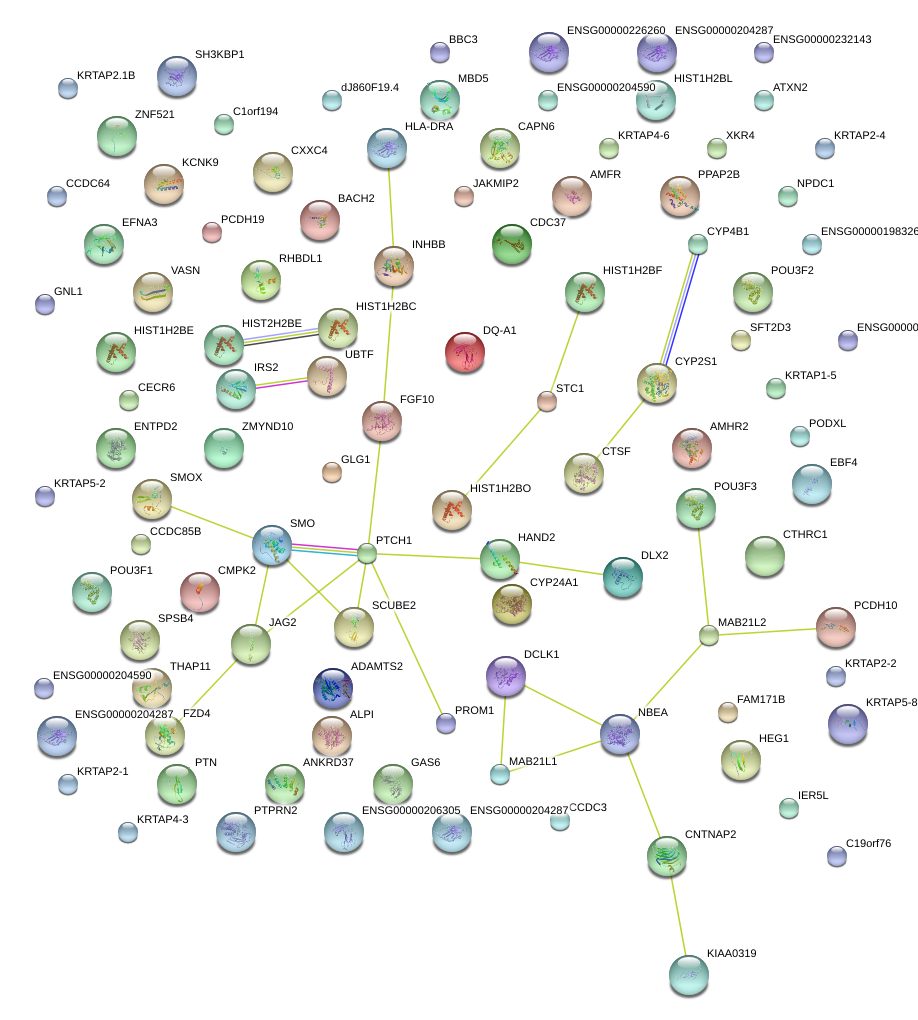
|
chr8_-_140715234
|
1.115
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr4_+_134073180
|
1.008
|
|
PCDH10
|
protocadherin 10
|
|
chr4_-_16077740
|
0.985
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr19_-_47734215
|
0.975
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr5_-_44388666
|
0.969
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr6_-_91006460
|
0.934
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr8_-_23712297
|
0.888
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr17_-_39324423
|
0.868
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr9_-_131940481
|
0.861
|
|
IER5L
|
immediate early response 5-like
|
|
chr13_-_36050758
|
0.851
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr13_+_36050885
|
0.838
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr6_+_99282597
|
0.811
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr3_-_124774735
|
0.778
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr1_+_47264669
|
0.771
|
NM_000779
NM_001099772
|
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1
|
|
chr16_+_4430878
|
0.757
|
|
VASN
|
vasorin
|
|
chrX_-_99665270
|
0.743
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr10_-_13043570
|
0.742
|
NM_031455
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr16_-_74640997
|
0.677
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr20_+_2673193
|
0.668
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr4_-_16085309
|
0.660
|
|
PROM1
|
prominin 1
|
|
chrX_-_110513750
|
0.657
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr6_-_30523502
|
0.656
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr16_-_74640915
|
0.653
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr2_-_39187477
|
0.634
|
|
LOC375196
|
uncharacterized LOC375196
|
|
chr17_-_39183453
|
0.613
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr4_-_174450529
|
0.591
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr2_+_121103670
|
0.585
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr6_-_27775683
|
0.582
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr2_-_172967627
|
0.575
|
|
DLX2
|
distal-less homeobox 2
|
|
chr11_+_71249070
|
0.565
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr6_-_91006580
|
0.563
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr9_-_131940538
|
0.562
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr2_+_148778573
|
0.559
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr16_+_67876381
|
0.540
|
|
THAP11
|
THAP domain containing 11
|
|
chr7_+_145813452
|
0.538
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr7_-_158380360
|
0.534
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr2_-_7005764
|
0.530
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr13_-_114567011
|
0.524
|
|
GAS6
|
growth arrest-specific 6
|
|
chr11_-_9113092
|
0.515
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr7_+_128828712
|
0.512
|
NM_005631
|
SMO
|
smoothened, frizzled family receptor
|
|
chr11_-_86666089
|
0.507
|
|
FZD4
|
frizzled family receptor 4
|
|
chr1_-_57045214
|
0.501
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr14_-_105634708
|
0.497
|
|
JAG2
|
jagged 2
|
|
chr1_-_109656478
|
0.497
|
NM_001122961
|
C1orf194
|
chromosome 1 open reading frame 194
|
|
chr18_-_22931970
|
0.493
|
|
ZNF521
|
zinc finger protein 521
|
|
chr13_-_36705432
|
0.490
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr7_-_131241321
|
0.489
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr16_-_56459371
|
0.486
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chr9_-_139948448
|
0.481
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr13_-_114567040
|
0.468
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr2_+_187558654
|
0.466
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr13_-_114566951
|
0.465
|
|
GAS6
|
growth arrest-specific 6
|
|
chr12_-_112036257
|
0.462
|
|
ATXN2
|
ataxin 2
|
|
chr19_+_41699076
|
0.455
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr16_-_74641014
|
0.452
|
NM_001145666
NM_001145667
NM_012201
|
GLG1
|
golgi glycoprotein 1
|
|
chr17_-_39296702
|
0.447
|
NM_030976
|
KRTAP4-6
|
keratin associated protein 4-6
|
|
chr2_-_172967234
|
0.447
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr11_-_66335958
|
0.442
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr13_-_114567034
|
0.440
|
|
GAS6
|
growth arrest-specific 6
|
|
chr8_+_56015013
|
0.439
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr14_-_105635114
|
0.439
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr20_+_2795613
|
0.434
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr2_+_128458943
|
0.429
|
|
SFT2D3
|
SFT2 domain containing 3
|
|
chr12_+_120427476
|
0.428
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr17_-_42297037
|
0.422
|
NM_001076684
NM_014233
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr9_-_139940556
|
0.422
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr7_-_137028275
|
0.413
|
|
PTN
|
pleiotrophin
|
|
chr8_+_104383785
|
0.411
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr11_+_65658101
|
0.402
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr9_-_98270830
|
0.399
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr1_+_155051263
|
0.399
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr20_-_52790511
|
0.398
|
NM_000782
NM_001128915
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1
|
|
chr3_-_50383009
|
0.397
|
NM_015896
|
ZMYND10
|
zinc finger, MYND-type containing 10
|
|
chr22_-_17602142
|
0.396
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr13_-_110438896
|
0.391
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chrX_-_19905605
|
0.390
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_+_53818864
|
0.387
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr5_-_178772430
|
0.386
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr4_-_105412466
|
0.383
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr16_+_725679
|
0.381
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr17_-_39203423
|
0.381
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr5_-_147162251
|
0.374
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr18_-_22932109
|
0.371
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr3_-_45267068
|
0.369
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr6_+_32407618
|
0.369
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr4_+_186317964
|
0.369
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr5_-_147162175
|
0.367
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_+_140770412
|
0.367
|
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr2_+_233320832
|
0.366
|
NM_001631
|
ALPI
|
alkaline phosphatase, intestinal
|
|
chr19_+_50192440
|
0.365
|
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr6_+_26183957
|
0.360
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr6_+_26158332
|
0.359
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr22_-_30783301
|
0.357
|
NM_001017981
|
RNF215
|
ring finger protein 215
|
|
chr6_-_132272311
|
0.357
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr16_+_69726751
|
0.354
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr8_+_104383701
|
0.353
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr6_+_157099873
|
0.353
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr10_-_133109983
|
0.351
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr12_-_71003622
|
0.351
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr14_-_54423448
|
0.350
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr22_-_31503484
|
0.345
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr11_+_71710971
|
0.342
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr17_-_42296923
|
0.338
|
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr9_-_84304378
|
0.336
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr11_+_76813884
|
0.336
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr5_-_148033715
|
0.335
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr1_-_57044980
|
0.335
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_165555167
|
0.334
|
|
BCHE
|
butyrylcholinesterase
|
|
chr6_+_157099203
|
0.333
|
|
|
|
|
chr5_+_99871023
|
0.330
|
NM_198507
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr16_+_29823557
|
0.327
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr17_-_7518124
|
0.326
|
NM_004860
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr6_-_32908583
|
0.326
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr8_+_85095452
|
0.324
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr3_+_110790460
|
0.324
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr5_-_147162056
|
0.322
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr20_+_19193364
|
0.322
|
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr18_+_77659446
|
0.320
|
|
|
|
|
chr9_-_139940464
|
0.320
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr5_-_138725544
|
0.318
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr1_-_57045129
|
0.317
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_4508955
|
0.316
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr2_-_230579198
|
0.316
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr2_-_190445466
|
0.315
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr19_+_3094528
|
0.315
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr19_+_18284631
|
0.311
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr2_+_115199898
|
0.311
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr11_+_57365577
|
0.307
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr19_+_41103055
|
0.306
|
NM_001042544
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr2_-_190445521
|
0.306
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr13_+_78271820
|
0.305
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr6_+_31973358
|
0.305
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr1_-_154842725
|
0.305
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_-_67390413
|
0.304
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr20_+_62368035
|
0.303
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr4_+_7194317
|
0.302
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr18_-_500583
|
0.300
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr22_-_18923654
|
0.300
|
|
PRODH
|
proline dehydrogenase (oxidase) 1
|
|
chr1_-_57045235
|
0.297
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr11_-_106888829
|
0.297
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr2_-_220436010
|
0.292
|
|
OBSL1
|
obscurin-like 1
|
|
chr16_-_75590107
|
0.290
|
NM_001077416
NM_001077418
NM_001077419
|
TMEM231
|
transmembrane protein 231
|
|
chr11_+_61976139
|
0.289
|
NM_002407
|
SCGB2A1
|
secretoglobin, family 2A, member 1
|
|
chrX_-_47489243
|
0.288
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr2_-_190445612
|
0.288
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chrX_-_140271275
|
0.288
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr9_-_139940595
|
0.288
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_+_218519576
|
0.287
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chrX_-_38079980
|
0.285
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr16_+_2802683
|
0.284
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr9_+_137533713
|
0.283
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr2_+_79740059
|
0.282
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr19_-_12845539
|
0.281
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr20_+_62367945
|
0.280
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr8_-_33457491
|
0.279
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr15_-_61521416
|
0.278
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr4_+_3768104
|
0.278
|
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr9_-_35812143
|
0.278
|
NM_001039592
NM_172312
|
SPAG8
|
sperm associated antigen 8
|
|
chr11_-_33891361
|
0.277
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr6_+_157099041
|
0.277
|
NM_017519
NM_020732
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr8_+_65493610
|
0.276
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr19_+_41699140
|
0.274
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr16_-_67427335
|
0.272
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr6_+_73331992
|
0.272
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr15_-_56035176
|
0.271
|
NM_173814
|
PRTG
|
protogenin
|
|
chr3_-_165555252
|
0.270
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr15_-_27018216
|
0.268
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chrX_-_140271293
|
0.267
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr20_+_56725982
|
0.267
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr7_-_45960738
|
0.266
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr12_+_26348488
|
0.266
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_-_42296865
|
0.266
|
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr11_-_33891485
|
0.264
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_-_23066828
|
0.264
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr9_-_34381497
|
0.263
|
NM_001252195
NM_147168
NM_147169
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr18_+_56530677
|
0.263
|
|
ZNF532
|
zinc finger protein 532
|
|
chr4_+_89299874
|
0.263
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr15_+_96873845
|
0.262
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr12_-_112037024
|
0.260
|
|
ATXN2
|
ataxin 2
|
|
chr19_-_7701852
|
0.259
|
|
|
|
|
chr19_+_18284578
|
0.259
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chrX_-_128788913
|
0.259
|
NM_017413
|
APLN
|
apelin
|
|
chr10_+_12391728
|
0.255
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr11_+_64001974
|
0.254
|
NM_001243733
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr1_-_58716173
|
0.254
|
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr18_-_46477080
|
0.253
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr11_-_33891258
|
0.253
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr2_-_220436267
|
0.252
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr15_-_37391596
|
0.251
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr4_+_89300110
|
0.251
|
|
HERC6
|
hect domain and RLD 6
|
|
chr4_+_15005364
|
0.250
|
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr6_-_30523933
|
0.250
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chrX_-_103087106
|
0.246
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr7_+_153749740
|
0.245
|
NM_130797
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr6_-_111136330
|
0.243
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_-_54519050
|
0.243
|
NM_004872
|
TMEM59
|
transmembrane protein 59
|
|
chr16_+_2802634
|
0.243
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr6_-_111136407
|
0.242
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chrX_-_19905665
|
0.242
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_54518790
|
0.241
|
|
TMEM59
|
transmembrane protein 59
|