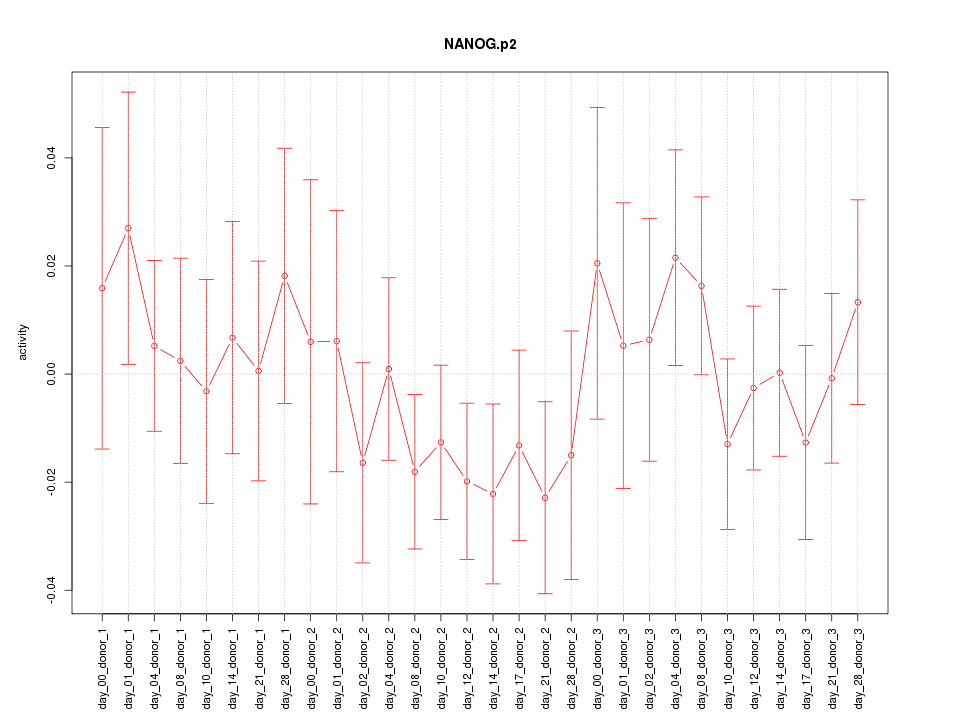
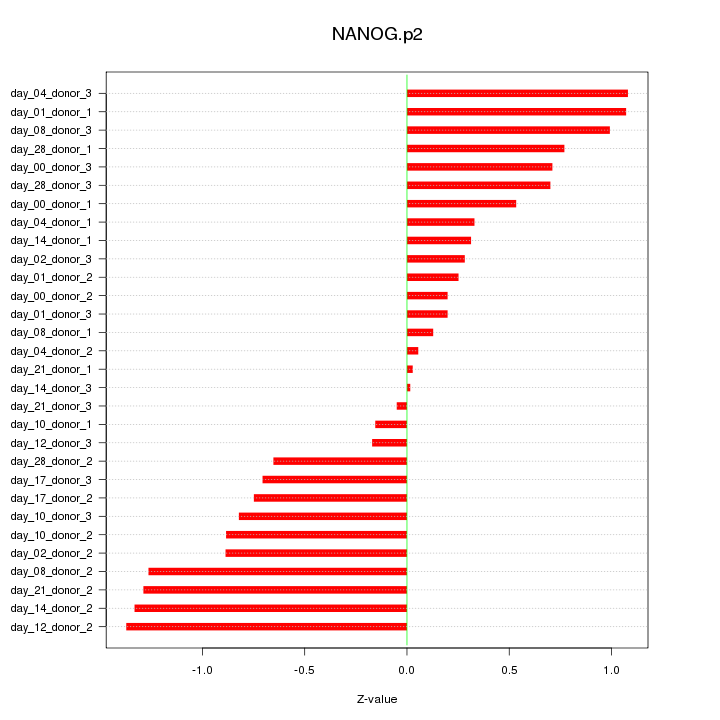
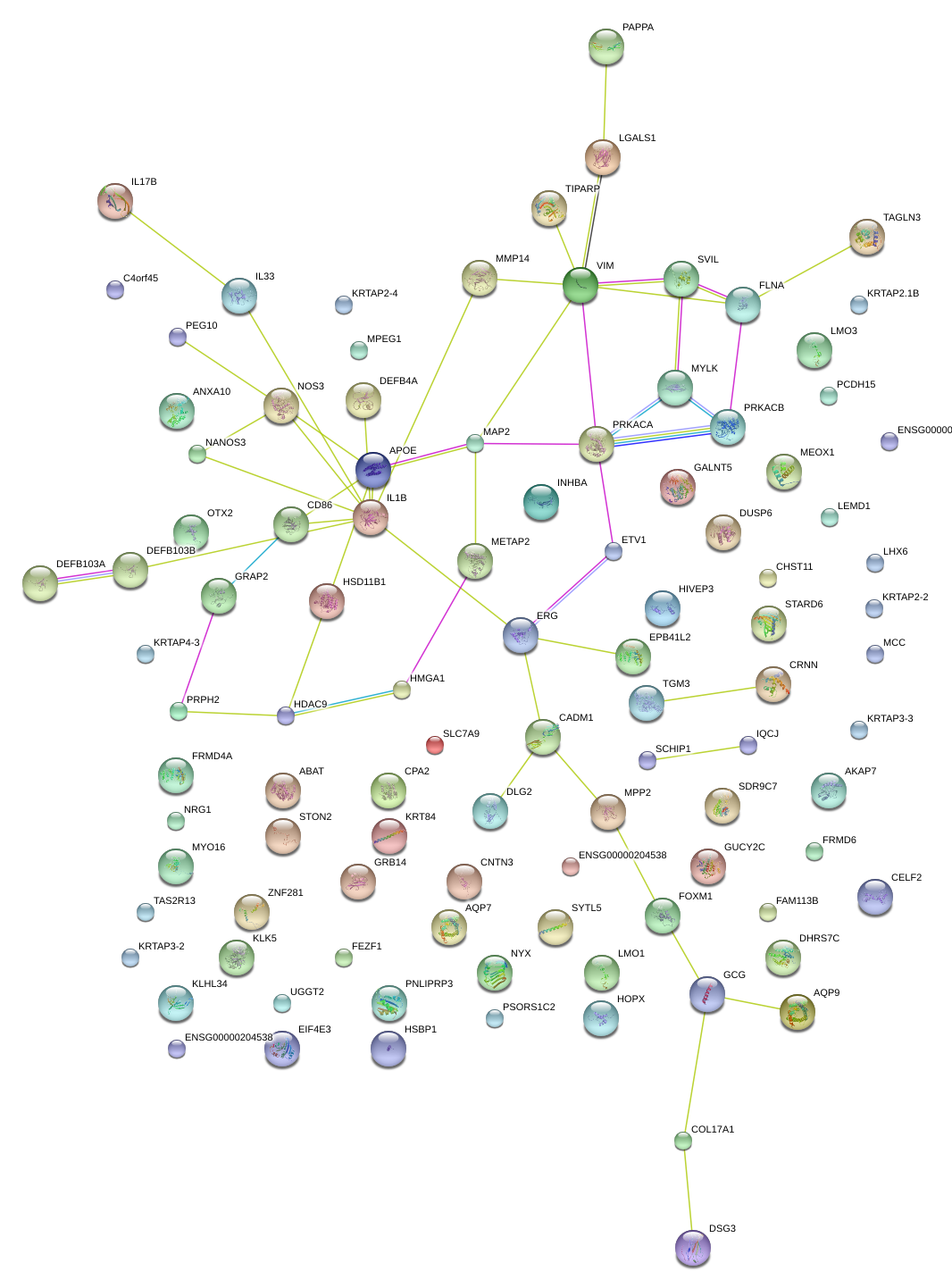
|
chr6_+_34206567
|
2.110
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_152386727
|
1.706
|
NM_016190
|
CRNN
|
cornulin
|
|
chr7_+_94297448
|
1.303
|
|
PEG10
|
paternally expressed 10
|
|
chr12_-_52779362
|
1.239
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr3_-_74570262
|
1.234
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr1_+_209878135
|
1.233
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr13_+_109281569
|
1.201
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr6_+_34204647
|
1.173
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_+_158114109
|
1.149
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr4_-_57547845
|
1.139
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr3_+_159557649
|
1.106
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_+_34204928
|
1.102
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_+_118187381
|
1.072
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr2_+_210444154
|
1.040
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_+_159557745
|
1.031
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chrX_+_37892786
|
1.018
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr2_+_210444364
|
0.970
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr6_+_34204690
|
0.936
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr9_-_124984018
|
0.898
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr6_-_131291480
|
0.861
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr12_-_89745997
|
0.859
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr9_+_6215785
|
0.851
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr4_+_169013687
|
0.839
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr11_-_115088628
|
0.781
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr12_-_89746244
|
0.768
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_-_9694613
|
0.764
|
NM_001105571
NM_001220493
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr6_+_34205029
|
0.753
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_-_113594325
|
0.753
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr10_-_105845619
|
0.752
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr10_-_105845556
|
0.744
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr3_-_71778268
|
0.737
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr9_-_124989755
|
0.728
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr12_-_89746238
|
0.723
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr7_-_14025826
|
0.711
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr22_+_38071612
|
0.704
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr12_-_16760935
|
0.688
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_41742666
|
0.684
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_+_121796695
|
0.660
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr7_+_18535351
|
0.626
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr10_-_14372807
|
0.617
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_102123420
|
0.595
|
|
|
|
|
chr5_-_148758787
|
0.588
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr10_-_56560947
|
0.587
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr5_-_112630301
|
0.570
|
|
MCC
|
mutated in colorectal cancers
|
|
chr3_+_111717585
|
0.558
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr8_-_7287651
|
0.553
|
NM_018661
|
DEFB103B
|
defensin, beta 103B
|
|
chr8_+_32504250
|
0.550
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr14_+_23305816
|
0.531
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr12_+_47610244
|
0.530
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr10_+_11206928
|
0.516
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr8_+_32579350
|
0.499
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr14_-_57272344
|
0.499
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr12_-_16759939
|
0.493
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_112630557
|
0.488
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr14_-_81864726
|
0.488
|
NM_033104
|
STON2
|
stonin 2
|
|
chr16_+_8814567
|
0.486
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_+_105153258
|
0.483
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_-_57328044
|
0.482
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr7_+_129906702
|
0.473
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr10_+_17270257
|
0.470
|
NM_003380
|
VIM
|
vimentin
|
|
chr1_+_84647342
|
0.470
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_+_150690838
|
0.466
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr1_-_42384370
|
0.464
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr18_-_51880942
|
0.454
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr6_+_34204663
|
0.454
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_-_165424993
|
0.454
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr10_-_29923900
|
0.449
|
NM_021738
|
SVIL
|
supervillin
|
|
chr8_-_7274353
|
0.448
|
NM_001205266
|
DEFB4B
|
defensin, beta 4B
|
|
chr12_-_16759733
|
0.434
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_+_51955854
|
0.431
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr1_-_205391180
|
0.430
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr20_+_2276612
|
0.428
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_-_39150384
|
0.426
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr3_-_123339423
|
0.420
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr6_+_43968303
|
0.420
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr7_-_121944485
|
0.414
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr6_-_31107110
|
0.412
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr6_+_34204576
|
0.405
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_-_39324423
|
0.403
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr18_+_29027731
|
0.402
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr10_+_11207053
|
0.401
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_+_156394452
|
0.399
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr14_+_23305792
|
0.385
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr1_-_200377488
|
0.382
|
|
ZNF281
|
zinc finger protein 281
|
|
chr13_-_60325073
|
0.381
|
|
|
|
|
chrX_-_21676416
|
0.378
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr11_-_83393436
|
0.377
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr4_-_159956332
|
0.371
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr7_-_14029641
|
0.368
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr17_-_41739261
|
0.367
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr19_-_33360641
|
0.365
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chrX_-_153602928
|
0.360
|
|
FLNA
|
filamin A, alpha
|
|
chr17_-_39211462
|
0.359
|
NM_033032
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr21_-_39956823
|
0.356
|
NM_001243429
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chrX_+_41306686
|
0.346
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr19_-_51456159
|
0.338
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr2_-_163008913
|
0.335
|
NM_002054
|
GCG
|
glucagon
|
|
chr12_+_47610051
|
0.335
|
NM_138371
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr9_+_118916069
|
0.335
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr3_-_123339178
|
0.330
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_14849427
|
0.330
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr19_+_45409003
|
0.330
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr15_+_58430394
|
0.327
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr11_-_58980321
|
0.322
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr22_+_40322609
|
0.321
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr6_+_131571298
|
0.317
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr12_-_11062160
|
0.300
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr4_+_71587674
|
0.296
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr19_-_14785626
|
0.291
|
NM_032571
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr8_+_26150731
|
0.288
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr16_+_87984948
|
0.283
|
NM_001173539
NM_001173540
NM_001173541
NM_017869
NM_079837
|
BANP
|
BTG3 associated nuclear protein
|
|
chr17_-_41276112
|
0.281
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr2_-_180427314
|
0.281
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr12_-_10021932
|
0.280
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr5_+_92918924
|
0.279
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr12_-_14132920
|
0.278
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr10_-_90712510
|
0.276
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr5_+_31193761
|
0.275
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr11_+_64018991
|
0.275
|
NM_000932
NM_001184883
|
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific)
|
|
chr1_+_16090993
|
0.273
|
NM_001024215
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr12_-_24103842
|
0.273
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr19_-_51522953
|
0.268
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr12_+_15475190
|
0.267
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr4_-_26491888
|
0.266
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr8_+_54764367
|
0.265
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr7_+_28725597
|
0.264
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_+_50574601
|
0.256
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_10022457
|
0.256
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr7_+_94285620
|
0.256
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr19_+_48325098
|
0.254
|
NM_000554
|
CRX
|
cone-rod homeobox
|
|
chr7_+_154002194
|
0.249
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr3_+_152880036
|
0.249
|
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr12_+_81110689
|
0.247
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr12_-_91505215
|
0.242
|
|
LUM
|
lumican
|
|
chr14_+_78870092
|
0.242
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr9_-_107690526
|
0.241
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr3_+_152880024
|
0.241
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr10_+_82173574
|
0.240
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr17_+_4854383
|
0.239
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr1_+_198608216
|
0.234
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr19_+_7030593
|
0.234
|
NM_001136507
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5
|
|
chrX_-_130423199
|
0.234
|
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr19_-_39235113
|
0.233
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr2_-_65593763
|
0.231
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr4_-_139163222
|
0.231
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr15_-_70390229
|
0.230
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr20_+_43030005
|
0.228
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_+_42295747
|
0.227
|
|
MYBL2
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 2
|
|
chr20_+_44441334
|
0.227
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr7_+_129984629
|
0.226
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr5_+_126984712
|
0.224
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr10_-_52645384
|
0.224
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr3_+_102153858
|
0.223
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr12_-_16757944
|
0.220
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_88529680
|
0.219
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr1_+_84609941
|
0.219
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_55199894
|
0.217
|
|
|
|
|
chr17_+_7905911
|
0.217
|
NM_000180
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific)
|
|
chr2_-_69098502
|
0.217
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr1_+_179560747
|
0.216
|
NM_001199091
NM_001199085
NM_173533
|
TDRD5
|
tudor domain containing 5
|
|
chr11_-_30601919
|
0.214
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr1_-_110155672
|
0.213
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr4_-_40936640
|
0.212
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr1_+_20617411
|
0.211
|
NM_001039500
|
VWA5B1
|
von Willebrand factor A domain containing 5B1
|
|
chr14_+_76837689
|
0.211
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chrX_+_28605558
|
0.208
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr4_-_69111333
|
0.206
|
NM_182502
|
TMPRSS11B
|
transmembrane protease, serine 11B
|
|
chr20_+_3776385
|
0.205
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr5_+_126988706
|
0.204
|
NM_001127385
|
CTXN3
|
cortexin 3
|
|
chr11_+_131240370
|
0.203
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr13_+_109248499
|
0.202
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr11_-_69634191
|
0.201
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr6_+_158438079
|
0.201
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chr4_-_80329354
|
0.201
|
NM_033214
|
GK2
|
glycerol kinase 2
|
|
chr1_-_178840079
|
0.200
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr7_+_28475233
|
0.199
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_-_111927059
|
0.199
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr12_-_4554779
|
0.199
|
NM_020996
|
FGF6
|
fibroblast growth factor 6
|
|
chr16_+_24740945
|
0.199
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr12_-_102874340
|
0.197
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr20_+_44441247
|
0.194
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr7_-_37025696
|
0.194
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_-_41750936
|
0.194
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr8_+_55528626
|
0.194
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr8_+_32785485
|
0.193
|
|
|
|
|
chr1_+_35246789
|
0.193
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr9_+_12693385
|
0.190
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr19_+_45971249
|
0.189
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_-_16759531
|
0.188
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_33172368
|
0.187
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_+_22050920
|
0.185
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr1_-_116383692
|
0.185
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr3_-_123411185
|
0.184
|
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_128294928
|
0.183
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr17_-_10452939
|
0.183
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr15_+_49715374
|
0.182
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr18_-_28742744
|
0.182
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr12_-_52828105
|
0.182
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr9_+_116263706
|
0.178
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_+_102508377
|
0.178
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|