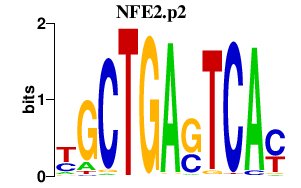
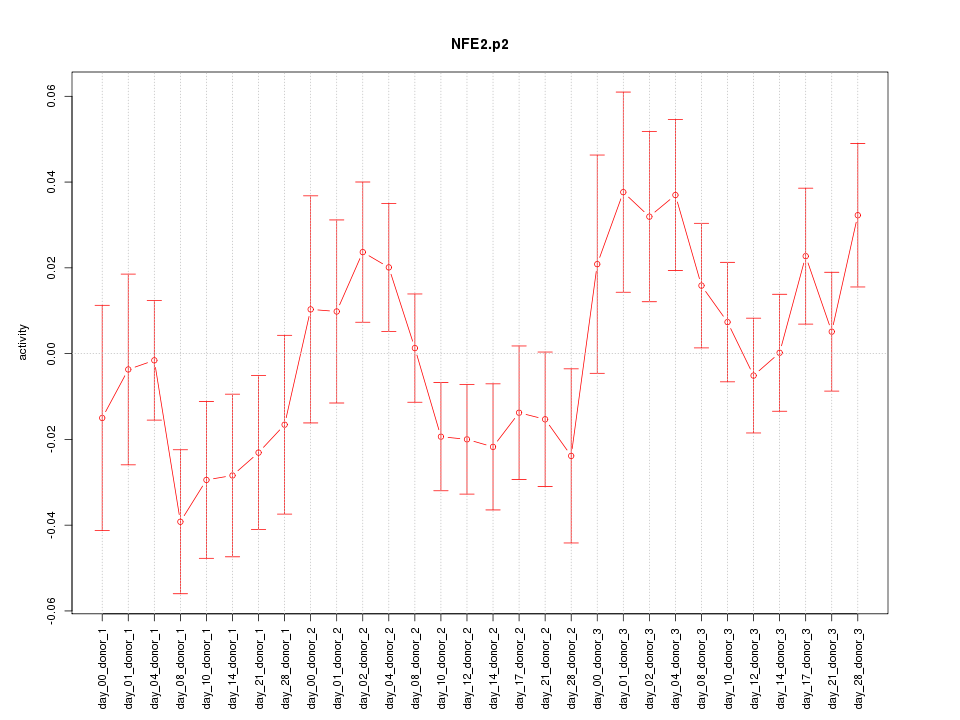
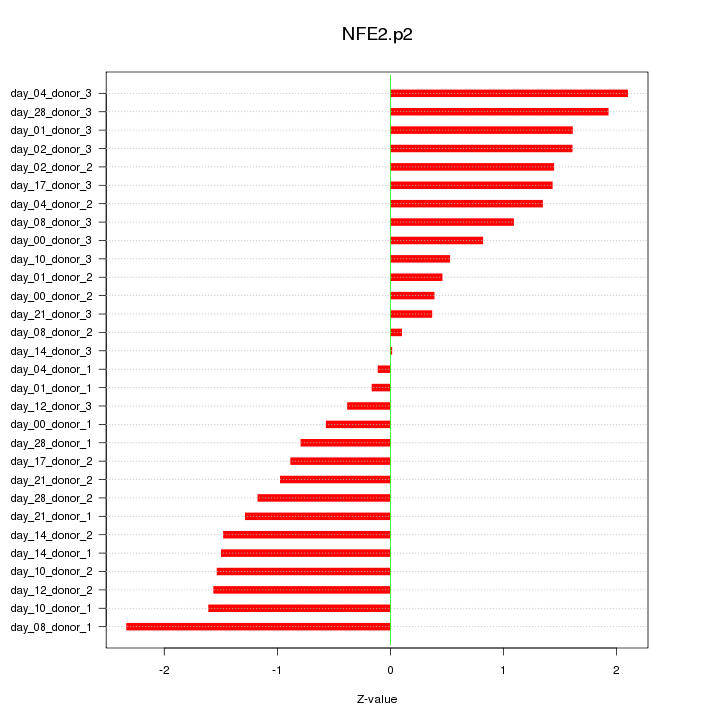
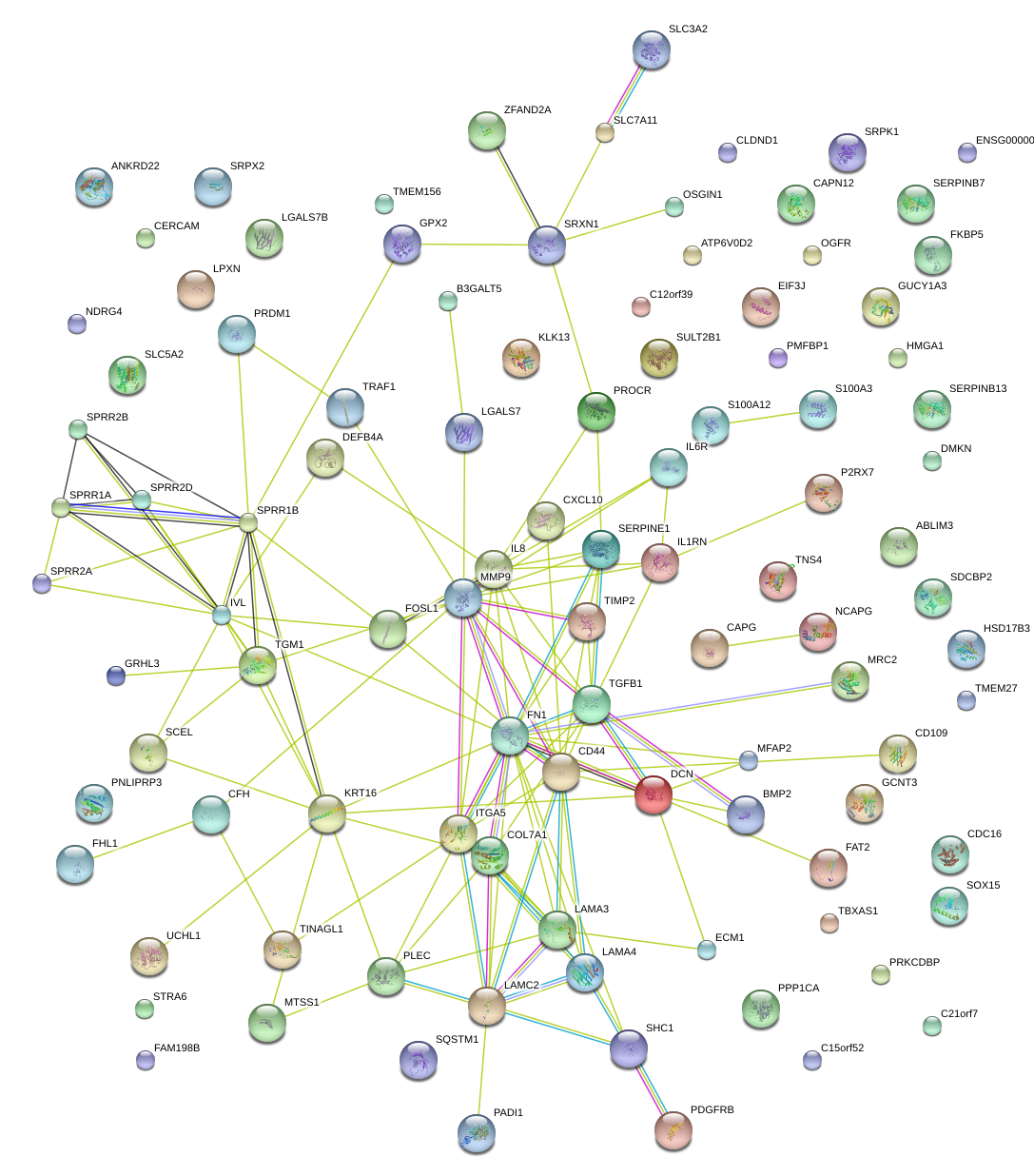
|
chr1_+_150480584
|
7.037
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr1_+_150480486
|
6.974
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr18_+_21452981
|
6.685
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr17_-_39768939
|
5.442
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr1_+_152881022
|
4.915
|
NM_005547
|
IVL
|
involucrin
|
|
chr2_-_216259197
|
4.900
|
|
FN1
|
fibronectin 1
|
|
chr21_+_41029253
|
4.356
|
NM_006057
NM_033170
NM_033171
NM_033172
NM_033173
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5
|
|
chr8_+_7752198
|
3.838
|
NM_004942
|
DEFB4A
|
defensin, beta 4A
|
|
chr1_+_183155173
|
3.794
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr2_+_113875469
|
3.670
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr11_-_65667858
|
3.598
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_153029987
|
3.491
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr1_-_153348057
|
3.485
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr1_+_32043745
|
3.471
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr8_-_7274353
|
3.410
|
NM_001205266
|
DEFB4B
|
defensin, beta 4B
|
|
chr6_+_74405764
|
3.335
|
|
CD109
|
CD109 molecule
|
|
chr1_+_183155393
|
3.052
|
|
LAMC2
|
laminin, gamma 2
|
|
chr14_-_24732344
|
3.042
|
NM_000359
|
TGM1
|
transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase)
|
|
chr6_+_74405505
|
2.899
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr1_+_17531619
|
2.880
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr1_+_183155361
|
2.872
|
|
LAMC2
|
laminin, gamma 2
|
|
chrX_+_99899162
|
2.849
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr17_-_76921205
|
2.817
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_+_74405933
|
2.770
|
|
CD109
|
CD109 molecule
|
|
chr11_-_65667860
|
2.742
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chrX_+_99899388
|
2.737
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr6_+_74405909
|
2.725
|
|
CD109
|
CD109 molecule
|
|
chr11_-_65667809
|
2.612
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr4_-_159093523
|
2.605
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_-_76921453
|
2.398
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_-_35696359
|
2.388
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr5_+_148520985
|
2.335
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr4_-_159092509
|
2.305
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr18_+_61442611
|
2.277
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr18_+_61254533
|
2.252
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr13_+_78109808
|
2.220
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr7_-_1199822
|
2.169
|
NM_182491
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr19_-_36019194
|
2.152
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr21_+_30517897
|
2.067
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr10_-_90611675
|
2.033
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr7_-_1199813
|
1.932
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr7_-_1199785
|
1.918
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr1_-_154943001
|
1.898
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr6_+_74405856
|
1.885
|
|
CD109
|
CD109 molecule
|
|
chr11_+_62649160
|
1.849
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr3_-_48632592
|
1.764
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr1_-_153013593
|
1.750
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr6_+_34206567
|
1.716
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_152956563
|
1.693
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr16_-_72206348
|
1.689
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr4_-_39033981
|
1.666
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr1_+_154377847
|
1.639
|
|
IL6R
|
interleukin 6 receptor
|
|
chr1_+_24649529
|
1.624
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr4_-_76944585
|
1.611
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr4_-_159093717
|
1.602
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr2_-_85641128
|
1.591
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr2_-_85641153
|
1.564
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr15_-_74494851
|
1.557
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr15_-_40630272
|
1.554
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr15_-_74494780
|
1.531
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr5_-_149535309
|
1.499
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr20_-_633844
|
1.495
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr19_-_39264144
|
1.480
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr6_+_106546736
|
1.443
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_+_39279849
|
1.430
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr19_-_39235113
|
1.428
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr20_+_44637539
|
1.420
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr1_+_154377668
|
1.391
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr12_-_54813002
|
1.376
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr16_+_58534045
|
1.355
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr12_-_91573264
|
1.354
|
NM_133503
|
DCN
|
decorin
|
|
chr1_+_154378044
|
1.342
|
|
IL6R
|
interleukin 6 receptor
|
|
chr1_-_17308072
|
1.340
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr17_+_60704747
|
1.336
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr7_+_100770378
|
1.334
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr20_-_1309809
|
1.324
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr20_+_33759773
|
1.321
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr1_-_153521717
|
1.271
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr19_-_51568323
|
1.269
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr16_-_72206023
|
1.265
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chrX_+_135278912
|
1.239
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_+_179247841
|
1.225
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr11_-_6341708
|
1.221
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr19_-_36004553
|
1.219
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chr3_-_98241357
|
1.196
|
|
CLDND1
|
claudin domain containing 1
|
|
chr8_-_125580745
|
1.196
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr4_+_41258941
|
1.196
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_74606222
|
1.185
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr10_+_118187381
|
1.165
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr4_-_139163222
|
1.144
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr19_-_41859253
|
1.143
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr14_-_65409445
|
1.134
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr17_-_38657814
|
1.091
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr2_+_87755059
|
1.086
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr8_-_145025043
|
1.068
|
NM_201380
|
PLEC
|
plectin
|
|
chr15_+_44829373
|
1.049
|
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr4_+_156587861
|
1.038
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr3_-_98241683
|
1.028
|
|
CLDND1
|
claudin domain containing 1
|
|
chr17_+_60705042
|
1.027
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr9_-_123688352
|
1.021
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr17_-_7493389
|
1.020
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr6_-_35888889
|
1.017
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr11_-_58343308
|
1.017
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr19_+_49055421
|
1.003
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr8_+_87111132
|
0.999
|
NM_152565
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2
|
|
chr16_+_31494438
|
0.997
|
NM_003041
|
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2
|
|
chrX_-_15683153
|
0.991
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr20_+_6748744
|
0.991
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr2_+_87754991
|
0.987
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr7_+_139528951
|
0.984
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr9_+_131191206
|
0.974
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr12_+_21679190
|
0.967
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr15_+_59904090
|
0.965
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr11_+_35160385
|
0.964
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
NM_001202555
NM_001202556
NM_001202557
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr16_+_83986826
|
0.961
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr9_-_99064385
|
0.958
|
NM_000197
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr15_+_59903980
|
0.957
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr5_-_149535138
|
0.943
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chrX_-_15511710
|
0.940
|
NM_001018109
NM_003662
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr7_+_143058794
|
0.940
|
|
ZYX
|
zyxin
|
|
chrX_-_106960268
|
0.918
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_+_60705051
|
0.913
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr12_+_56522043
|
0.905
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr1_+_35246789
|
0.901
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr12_+_56521953
|
0.899
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr8_-_145016691
|
0.898
|
NM_201383
|
PLEC
|
plectin
|
|
chr9_+_4985082
|
0.886
|
|
JAK2
|
Janus kinase 2
|
|
chr2_-_172016904
|
0.882
|
|
TLK1
|
tousled-like kinase 1
|
|
chr2_+_28615771
|
0.881
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_+_9745847
|
0.876
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr3_-_98241748
|
0.870
|
|
CLDND1
|
claudin domain containing 1
|
|
chrX_-_153285262
|
0.865
|
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr6_-_35888816
|
0.858
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr19_-_44285012
|
0.852
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr5_-_175843531
|
0.849
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chr12_+_56522202
|
0.846
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr4_+_110834039
|
0.841
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr12_+_56522049
|
0.837
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr6_+_32121228
|
0.829
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr2_+_28615695
|
0.829
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr6_-_35888937
|
0.827
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr3_-_98241722
|
0.826
|
|
CLDND1
|
claudin domain containing 1
|
|
chr11_+_35160666
|
0.824
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr11_-_62314199
|
0.820
|
|
AHNAK
|
AHNAK nucleoprotein
|
|
chr5_-_149535420
|
0.820
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_54812936
|
0.817
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr2_+_33359607
|
0.815
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr9_-_35111534
|
0.803
|
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr14_+_79745653
|
0.797
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr7_+_143078444
|
0.796
|
|
ZYX
|
zyxin
|
|
chr9_+_140119643
|
0.793
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr7_+_143078447
|
0.786
|
|
ZYX
|
zyxin
|
|
chr2_-_190044330
|
0.785
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_+_32504250
|
0.784
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr8_-_143823823
|
0.783
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr20_-_633808
|
0.778
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr9_-_130341265
|
0.776
|
NM_001035534
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr6_+_64231626
|
0.765
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_+_40627066
|
0.758
|
|
RLF
|
rearranged L-myc fusion
|
|
chr19_+_56347943
|
0.757
|
NM_134444
|
NLRP4
|
NLR family, pyrin domain containing 4
|
|
chr3_+_160394946
|
0.756
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr12_-_49259549
|
0.755
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr1_+_84647342
|
0.750
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr14_-_25078818
|
0.748
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr10_+_17270257
|
0.745
|
NM_003380
|
VIM
|
vimentin
|
|
chr7_+_143078457
|
0.737
|
|
ZYX
|
zyxin
|
|
chr10_-_76868853
|
0.735
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr20_-_34195409
|
0.733
|
|
FER1L4
|
fer-1-like 4 (C. elegans) pseudogene
|
|
chr22_-_37584254
|
0.725
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr9_+_116356407
|
0.720
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr19_-_44285407
|
0.719
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr7_-_86688944
|
0.714
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr5_-_175843360
|
0.700
|
|
CLTB
|
clathrin, light chain B
|
|
chr19_-_51537996
|
0.697
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr2_-_65593866
|
0.693
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr17_+_74381186
|
0.691
|
NM_001142602
|
SPHK1
|
sphingosine kinase 1
|
|
chr19_-_41858948
|
0.691
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr11_+_128563993
|
0.685
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_+_36621751
|
0.680
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_+_4985243
|
0.671
|
NM_004972
|
JAK2
|
Janus kinase 2
|
|
chr7_-_93519990
|
0.670
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr7_+_134212343
|
0.669
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr2_-_69870705
|
0.668
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr12_-_48152860
|
0.651
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_87040246
|
0.648
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr7_+_143078358
|
0.641
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr7_+_143078399
|
0.636
|
|
ZYX
|
zyxin
|
|
chr21_-_35986744
|
0.634
|
NM_203417
|
RCAN1
|
regulator of calcineurin 1
|
|
chr3_+_49208979
|
0.634
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr1_-_24469571
|
0.634
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr12_-_53242716
|
0.632
|
NM_173352
|
KRT78
|
keratin 78
|
|
chr2_+_74057969
|
0.632
|
|
STAMBP
|
STAM binding protein
|
|
chr2_-_190927321
|
0.631
|
NM_005259
|
MSTN
|
myostatin
|
|
chr5_-_158636489
|
0.630
|
NM_001199383
|
RNF145
|
ring finger protein 145
|
|
chr4_+_77953942
|
0.628
|
|
SEPT11
|
septin 11
|
|
chr8_+_11946846
|
0.626
|
NM_001039615
|
ZNF705D
|
zinc finger protein 705D
|
|
chr4_-_80329354
|
0.625
|
NM_033214
|
GK2
|
glycerol kinase 2
|
|
chr19_+_6531009
|
0.623
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr16_+_56642477
|
0.621
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr12_-_10324675
|
0.620
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|