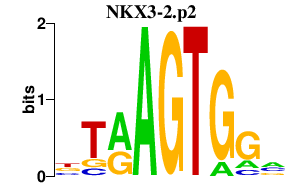
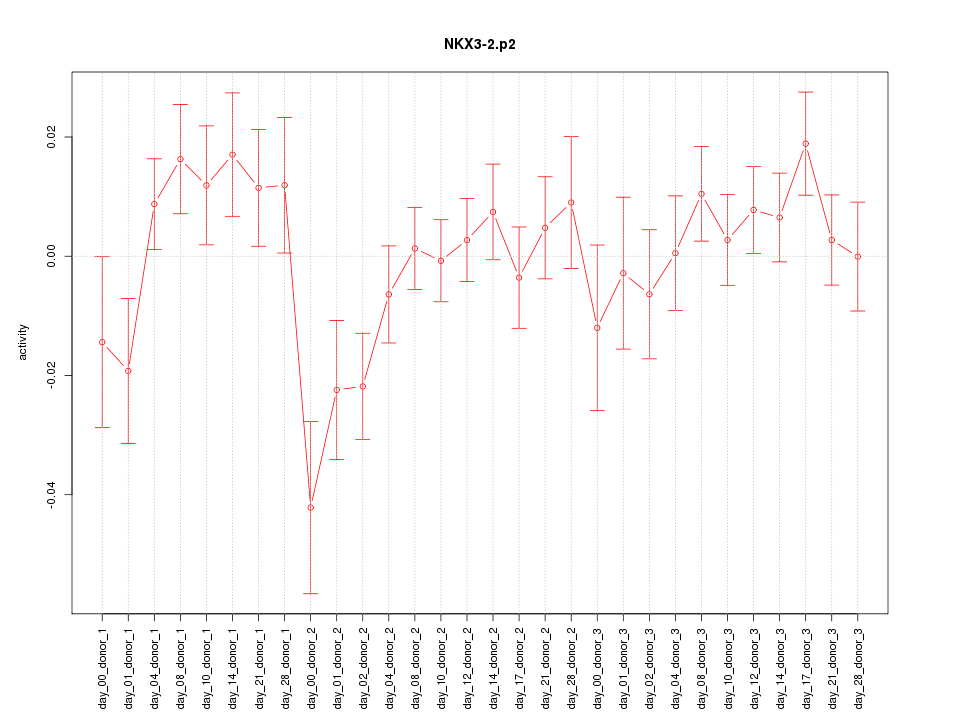
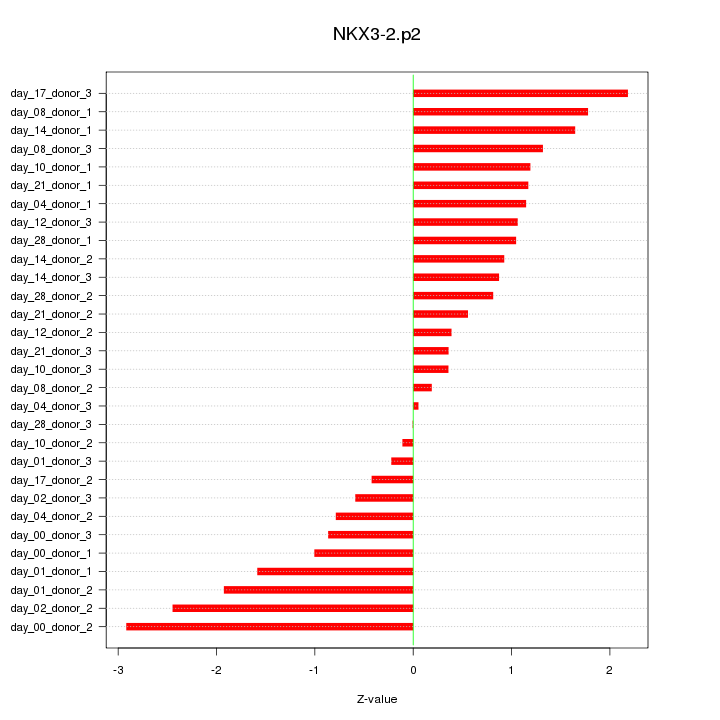
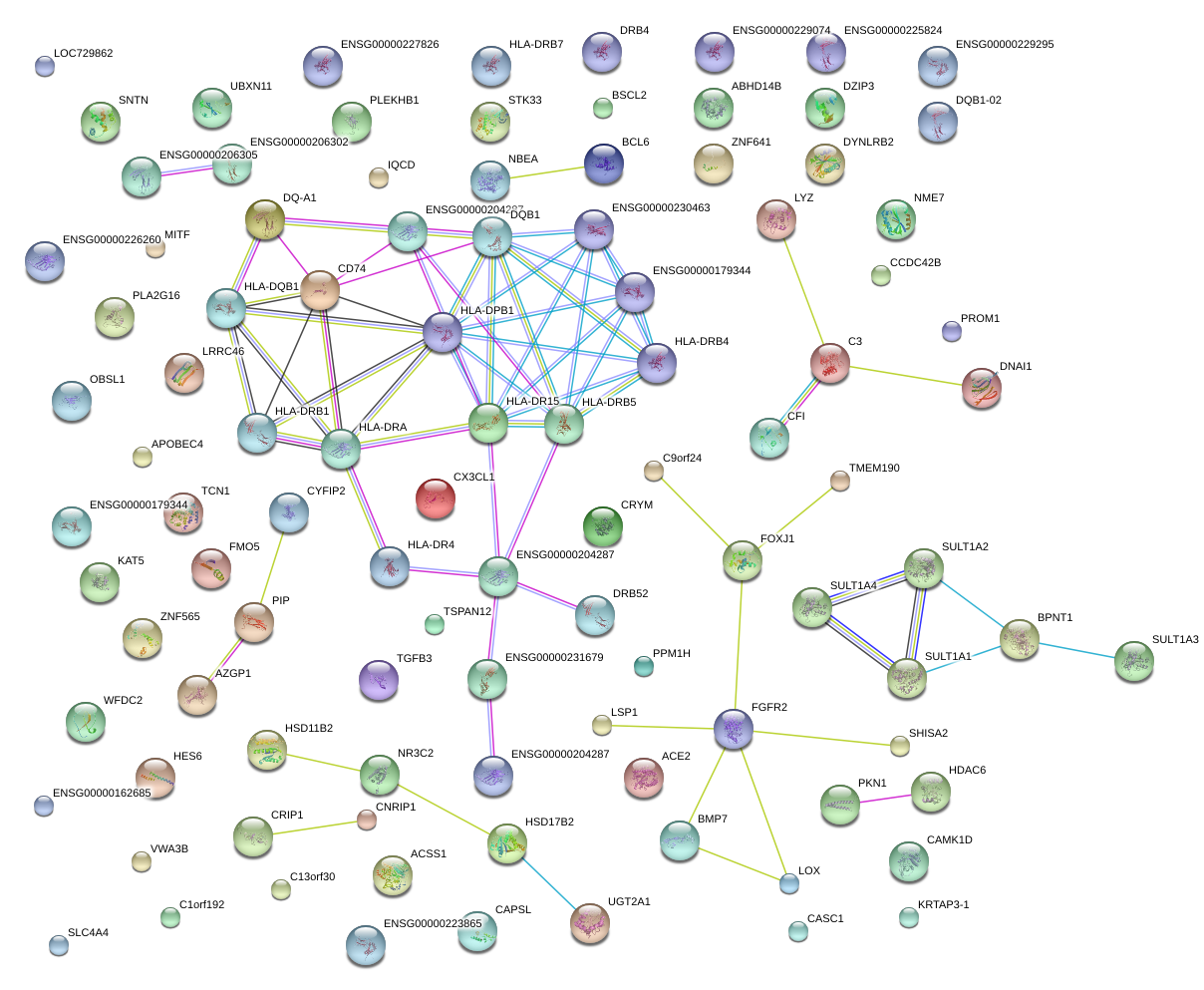
|
chr6_-_32497925
|
7.828
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr16_+_57406398
|
6.493
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_-_149792370
|
6.163
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr20_+_31870940
|
5.325
|
NM_033197
|
BPIFB1
|
BPI fold containing family B, member 1
|
|
chr4_-_16077740
|
5.024
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr9_-_34397786
|
5.021
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr17_-_74137370
|
4.964
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr1_-_161337663
|
4.847
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr6_+_32407618
|
4.756
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr6_-_32557431
|
3.994
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr4_-_110723127
|
3.838
|
|
CFI
|
complement factor I
|
|
chr5_-_149792276
|
3.835
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr4_-_110723334
|
3.694
|
NM_000204
|
CFI
|
complement factor I
|
|
chr9_+_34458810
|
3.604
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr4_-_149363498
|
3.602
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr12_-_25348031
|
3.503
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr5_-_35938736
|
3.393
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr11_+_65479685
|
3.313
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr16_-_21289637
|
3.114
|
|
CRYM
|
crystallin, mu
|
|
chr16_+_82068830
|
3.040
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr19_+_55888203
|
3.022
|
NM_139172
|
TMEM190
|
transmembrane protein 190
|
|
chr7_-_99573637
|
2.880
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr14_+_105953531
|
2.648
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr3_+_63638343
|
2.633
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr1_-_183622447
|
2.623
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr20_-_55841090
|
2.521
|
|
BMP7
|
bone morphogenetic protein 7
|
|
chr1_-_146697155
|
2.399
|
NM_001144829
NM_001144830
NM_001461
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr17_+_45908992
|
2.334
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr5_+_156696351
|
2.328
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr13_+_35516390
|
2.239
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr16_-_21289571
|
2.173
|
|
CRYM
|
crystallin, mu
|
|
chr19_-_6720585
|
2.161
|
NM_000064
|
C3
|
complement component 3
|
|
chr12_-_48744553
|
2.153
|
NM_001172681
NM_152320
|
ZNF641
|
zinc finger protein 641
|
|
chr11_-_62474810
|
2.135
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr2_-_239148545
|
2.078
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr16_+_80574853
|
2.046
|
NM_130897
|
DYNLRB2
|
dynein, light chain, roadblock-type 2
|
|
chr9_-_3223007
|
2.024
|
|
|
|
|
chr1_-_169336999
|
2.009
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr10_-_123353771
|
1.953
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr3_+_108308528
|
1.937
|
|
DZIP3
|
DAZ interacting protein 3, zinc finger
|
|
chr11_-_63381485
|
1.913
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr13_-_26625164
|
1.911
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr7_+_142829169
|
1.877
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr4_+_72052354
|
1.766
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr20_-_25038501
|
1.733
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr12_-_113658826
|
1.717
|
NM_138451
|
IQCD
|
IQ motif containing D
|
|
chr11_-_8615412
|
1.684
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr3_-_187454247
|
1.659
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr13_+_43355685
|
1.652
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr14_+_62584120
|
1.639
|
|
FLJ43390
|
uncharacterized LOC646113
|
|
chr16_+_67465019
|
1.628
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr10_+_12391728
|
1.622
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr2_-_220436267
|
1.620
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr12_+_69742130
|
1.612
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr19_+_14551080
|
1.591
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr19_-_36705982
|
1.587
|
NM_001042474
|
ZNF565
|
zinc finger protein 565
|
|
chr12_+_113587547
|
1.569
|
NM_001144872
|
CCDC42B
|
coiled-coil domain containing 42B
|
|
chr11_+_73359734
|
1.564
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr17_-_39165343
|
1.549
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr20_+_44098392
|
1.523
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr11_-_59633942
|
1.519
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr3_+_69915374
|
1.514
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_-_15620191
|
1.494
|
NM_021804
|
ACE2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr1_-_26644755
|
1.494
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr7_-_120498009
|
1.471
|
|
TSPAN12
|
tetraspanin 12
|
|
chr5_-_121412805
|
1.470
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr7_-_120498128
|
1.458
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr16_+_29471206
|
1.458
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr2_+_98703594
|
1.455
|
NM_144992
|
VWA3B
|
von Willebrand factor A domain containing 3B
|
|
chr12_-_63328663
|
1.453
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr10_+_12391708
|
1.439
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chrX_+_48660467
|
1.437
|
NM_006044
|
HDAC6
|
histone deacetylase 6
|
|
chr11_+_1889902
|
1.433
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr14_-_76448091
|
1.428
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr3_-_52008012
|
1.413
|
|
ABHD14B
|
abhydrolase domain containing 14B
|
|
chr11_-_8615745
|
1.404
|
|
STK33
|
serine/threonine kinase 33
|
|
chr4_-_70505359
|
1.389
|
NM_001105677
|
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2
|
|
chr1_-_217262946
|
1.388
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_+_48507621
|
1.384
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr3_+_48507209
|
1.380
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr17_+_68071386
|
1.379
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr21_+_42688660
|
1.374
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr6_-_79787887
|
1.373
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chrX_-_44202834
|
1.369
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr17_-_6735007
|
1.367
|
NM_053285
|
TEKT1
|
tektin 1
|
|
chr2_+_85766291
|
1.365
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr19_+_14551087
|
1.361
|
|
PKN1
|
protein kinase N1
|
|
chr1_+_118148506
|
1.356
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr2_-_220436010
|
1.348
|
|
OBSL1
|
obscurin-like 1
|
|
chr1_+_47264669
|
1.339
|
NM_000779
NM_001099772
|
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1
|
|
chr1_+_118148668
|
1.332
|
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr5_-_42811959
|
1.331
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr17_-_19281494
|
1.330
|
NM_001243475
|
B9D1
|
B9 protein domain 1
|
|
chr2_-_86564632
|
1.326
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr6_-_79787784
|
1.325
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr1_+_212738675
|
1.322
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_146696896
|
1.316
|
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr10_+_7745354
|
1.315
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr7_+_105603656
|
1.315
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr4_-_141348760
|
1.297
|
NM_001130675
NM_004362
|
CLGN
|
calmegin
|
|
chr20_-_21494663
|
1.292
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr4_+_55523906
|
1.280
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr3_+_133495925
|
1.280
|
|
TF
|
transferrin
|
|
chr2_+_114384811
|
1.277
|
NM_007082
NM_013412
|
RABL2A
|
RAB, member of RAS oncogene family-like 2A
|
|
chr12_+_41831484
|
1.276
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr17_+_4981754
|
1.256
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr10_+_7745229
|
1.256
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr19_-_1876133
|
1.254
|
|
|
|
|
chr10_+_7745325
|
1.254
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr11_-_8615701
|
1.243
|
|
STK33
|
serine/threonine kinase 33
|
|
chr14_-_54421269
|
1.242
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr16_-_28621279
|
1.233
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr19_+_14551052
|
1.232
|
|
PKN1
|
protein kinase N1
|
|
chr17_+_1633754
|
1.230
|
|
WDR81
|
WD repeat domain 81
|
|
chr7_-_131241321
|
1.221
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr2_-_25194772
|
1.214
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr4_+_41700736
|
1.213
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_-_28608367
|
1.210
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr2_+_106682111
|
1.207
|
NM_032411
|
C2orf40
|
chromosome 2 open reading frame 40
|
|
chr2_-_25016214
|
1.199
|
NM_001013663
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1
|
|
chr3_-_120400957
|
1.199
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr20_+_43990576
|
1.187
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr16_+_82068958
|
1.184
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr2_-_148779172
|
1.180
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr11_+_100557851
|
1.174
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr8_+_123793894
|
1.173
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr12_-_122107559
|
1.167
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr21_-_33984657
|
1.155
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr1_-_150738255
|
1.154
|
|
CTSS
|
cathepsin S
|
|
chrX_-_132548994
|
1.145
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr14_+_65016613
|
1.142
|
NM_172365
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36
|
|
chr1_+_32674694
|
1.132
|
NM_001099434
|
DCDC2B
|
doublecortin domain containing 2B
|
|
chr13_+_24144402
|
1.131
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr12_-_42983412
|
1.120
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr12_+_69742140
|
1.113
|
|
LYZ
|
lysozyme
|
|
chr16_-_4896197
|
1.113
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr1_+_46646199
|
1.107
|
|
TSPAN1
|
tetraspanin 1
|
|
chr8_+_18067614
|
1.105
|
NM_000662
NM_001160170
NM_001160171
NM_001160172
NM_001160173
NM_001160175
NM_001160176
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr12_+_20963637
|
1.104
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr21_-_33984865
|
1.102
|
NM_021254
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chrX_-_151999259
|
1.101
|
NM_004344
|
CETN2
|
centrin, EF-hand protein, 2
|
|
chr10_+_12391471
|
1.094
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr4_-_105412466
|
1.092
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr3_-_145968819
|
1.091
|
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr7_-_25164902
|
1.091
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr6_-_165723074
|
1.083
|
NM_144980
|
C6orf118
|
chromosome 6 open reading frame 118
|
|
chr9_-_75568232
|
1.075
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chrX_-_8700170
|
1.071
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr4_+_77356252
|
1.070
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr8_-_27115935
|
1.068
|
|
STMN4
|
stathmin-like 4
|
|
chr9_-_75567915
|
1.066
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr6_+_146920135
|
1.062
|
NM_024694
|
C6orf103
|
chromosome 6 open reading frame 103
|
|
chr17_+_4981721
|
1.059
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr19_+_16999807
|
1.059
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr1_+_20512577
|
1.053
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr3_-_120401401
|
1.048
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr3_-_169530306
|
1.037
|
|
LRRC34
|
leucine rich repeat containing 34
|
|
chr15_-_68498373
|
1.035
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr1_-_217263195
|
1.029
|
NM_001243505
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_+_123793988
|
1.024
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr6_-_46889620
|
1.012
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr4_-_17806820
|
1.006
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr5_+_35617945
|
1.005
|
NM_024867
NM_144722
|
SPEF2
|
sperm flagellar 2
|
|
chr4_+_81256873
|
1.000
|
NM_001206997
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chr8_-_110657745
|
0.996
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr7_-_158380360
|
0.995
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr2_+_190648810
|
0.991
|
NM_000534
NM_001128143
NM_001128144
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae)
|
|
chr10_-_116286570
|
0.988
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_158731622
|
0.987
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chrX_+_48660561
|
0.984
|
|
HDAC6
|
histone deacetylase 6
|
|
chr8_+_123794043
|
0.976
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr22_-_38349533
|
0.966
|
NM_032561
NM_001207062
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chrX_-_112084006
|
0.964
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr9_+_71320105
|
0.955
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr4_-_168155576
|
0.953
|
NM_001204353
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr1_-_75139421
|
0.950
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr6_-_52774460
|
0.949
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr3_-_114343791
|
0.947
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_122526410
|
0.940
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr13_-_31735718
|
0.940
|
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr22_-_31503484
|
0.936
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr19_+_36236460
|
0.933
|
NM_172341
|
PSENEN
|
presenilin enhancer 2 homolog (C. elegans)
|
|
chr6_-_31324955
|
0.928
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr3_-_182880463
|
0.923
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr20_+_18794369
|
0.918
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr5_+_42565963
|
0.917
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr3_-_145968958
|
0.915
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr7_+_30960914
|
0.912
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr7_+_90339146
|
0.902
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr2_+_39103102
|
0.901
|
NM_001145450
|
MORN2
|
MORN repeat containing 2
|
|
chr1_-_216978708
|
0.901
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr19_+_41284161
|
0.900
|
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 readthrough
RAB4B, member RAS oncogene family
|
|
chrX_+_48660286
|
0.890
|
|
HDAC6
|
histone deacetylase 6
|
|
chr5_-_41213513
|
0.888
|
NM_000065
|
C6
|
complement component 6
|
|
chr17_-_38721701
|
0.883
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr1_+_59775749
|
0.881
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr6_-_31324975
|
0.879
|
NM_005514
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr3_-_46506342
|
0.873
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr8_-_110704019
|
0.868
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_-_31324898
|
0.866
|
|
HLA-B
|
major histocompatibility complex, class I, B
|