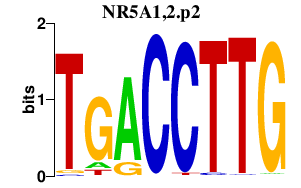
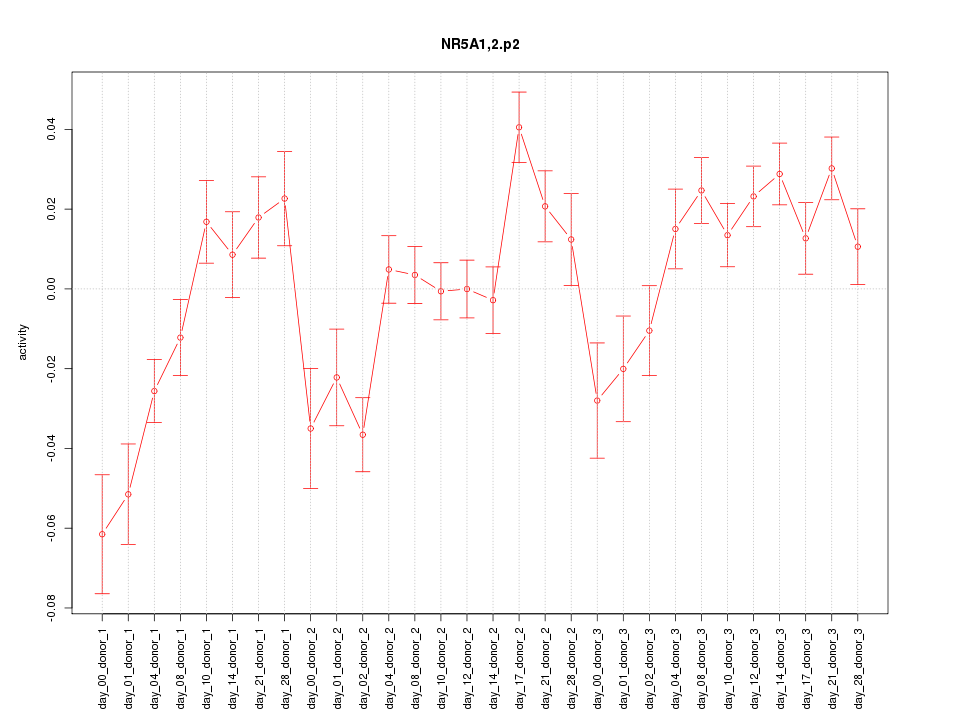
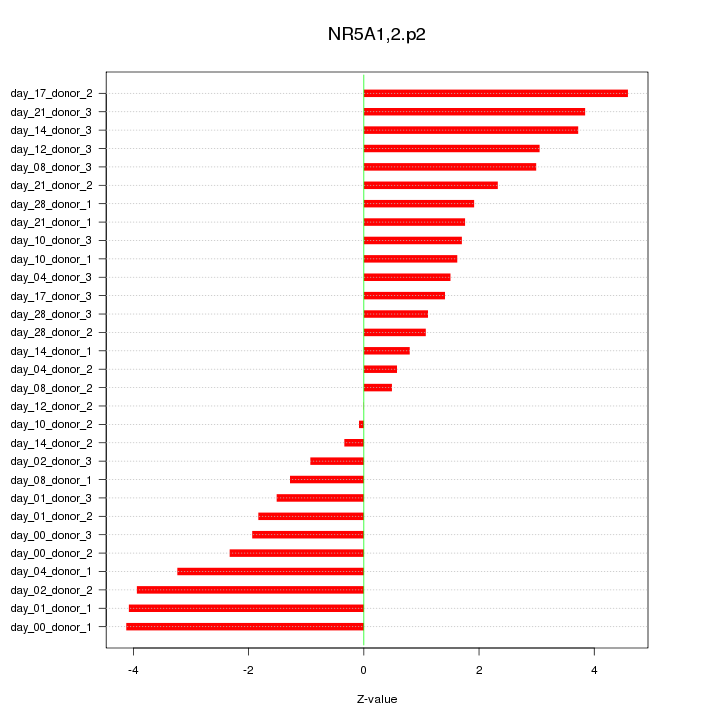
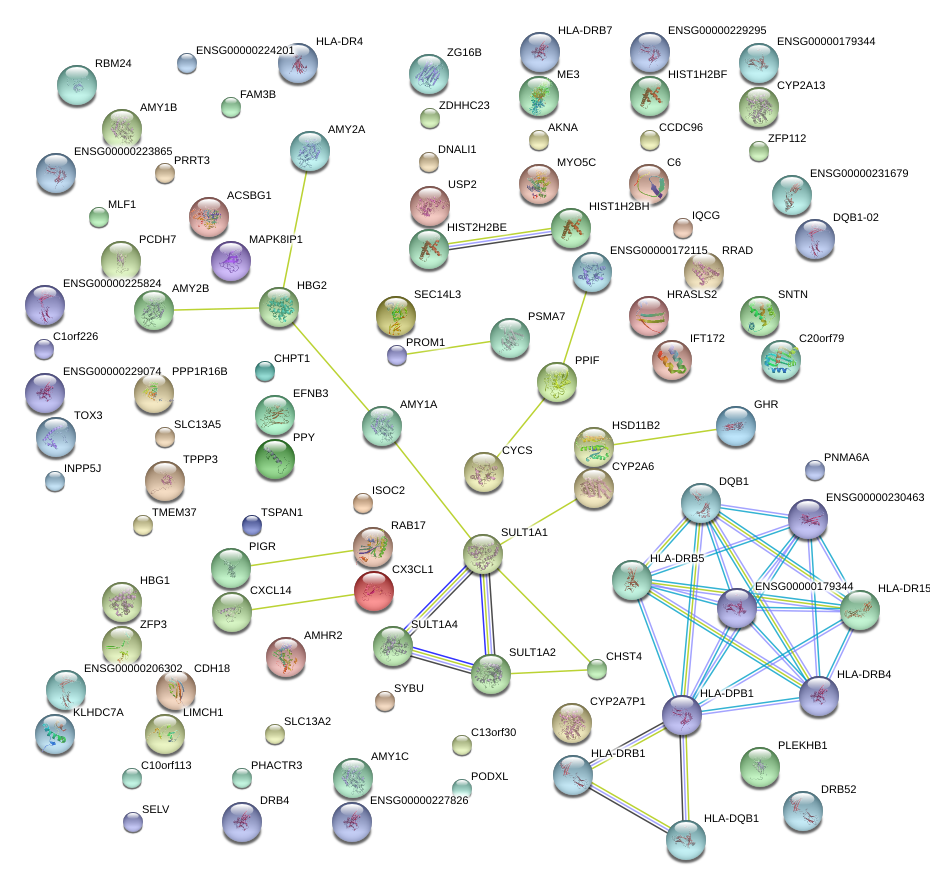
|
chr16_-_67427335
|
9.545
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr3_+_63638343
|
7.798
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr1_+_104198140
|
7.760
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr1_+_162351519
|
7.498
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr1_-_207119707
|
7.446
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr1_+_104198301
|
7.384
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr19_+_41594367
|
7.338
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr6_-_32551993
|
6.595
|
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr11_-_5275904
|
6.530
|
NM_000184
|
HBG1
HBG2
|
hemoglobin, gamma A
hemoglobin, gamma G
|
|
chr16_+_67465019
|
6.399
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr2_+_120189342
|
6.015
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr13_+_43355685
|
5.759
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr1_+_38022514
|
5.378
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr4_+_41614911
|
5.278
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr19_-_44831907
|
5.132
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr1_-_104239071
|
5.096
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr11_-_5271035
|
5.024
|
NM_000559
|
HBG1
|
hemoglobin, gamma A
|
|
chr4_-_7044664
|
4.850
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr3_-_197676755
|
4.843
|
NM_001134435
|
IQCG
|
IQ motif containing G
|
|
chr19_-_41356193
|
4.842
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr5_+_42565562
|
4.817
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr5_+_42565963
|
4.753
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr16_-_52580805
|
4.720
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr1_-_149858114
|
4.616
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr21_+_42688660
|
4.531
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr2_-_238499713
|
4.530
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr16_+_57406398
|
4.518
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr7_-_131241321
|
4.479
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr1_-_104238785
|
4.395
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr9_-_117150217
|
4.386
|
|
AKNA
|
AT-hook transcription factor
|
|
chr8_-_110657025
|
4.309
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr4_+_41614933
|
4.247
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_42019801
|
4.239
|
NM_002722
|
PPY
|
pancreatic polypeptide
|
|
chr4_+_30722191
|
4.229
|
|
PCDH7
|
protocadherin 7
|
|
chr2_-_238499317
|
4.047
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr16_-_66959418
|
4.034
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr11_+_73359734
|
3.913
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr17_+_26800657
|
3.864
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr12_+_102091399
|
3.816
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr11_-_86383164
|
3.777
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr5_-_41213513
|
3.639
|
NM_000065
|
C6
|
complement component 6
|
|
chr16_-_28608367
|
3.627
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr6_+_17282931
|
3.590
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr20_+_58152563
|
3.560
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr16_+_71560022
|
3.550
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr22_+_31518923
|
3.537
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr4_+_30721865
|
3.520
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr11_-_63330854
|
3.510
|
NM_017878
|
HRASLS2
|
HRAS-like suppressor 2
|
|
chr10_+_81107259
|
3.495
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr7_-_25164902
|
3.434
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr17_+_4981754
|
3.338
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr11_-_119234628
|
3.335
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr9_-_117156684
|
3.335
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr5_-_134914968
|
3.285
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr10_-_21435487
|
3.267
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr1_+_46640748
|
3.265
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr20_+_31870940
|
3.240
|
NM_033197
|
BPIFB1
|
BPI fold containing family B, member 1
|
|
chr10_+_81107242
|
3.230
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr2_-_27712520
|
3.168
|
NM_015662
|
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas)
|
|
chr17_+_7608413
|
3.162
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr3_-_9994057
|
3.161
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr1_+_18807423
|
3.136
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr19_-_55973048
|
3.121
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chrX_+_152240726
|
3.120
|
NM_001170944
NM_001242319
NM_032882
|
PNMA6C
PNMA6D
PNMA6A
|
paraneoplastic antigen like 6C
paraneoplastic antigen like 6D
paraneoplastic antigen like 6A
|
|
chr11_+_45907046
|
3.109
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr4_-_16077740
|
3.047
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr3_+_158288952
|
3.027
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr3_+_113666544
|
3.019
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr20_+_37434294
|
3.014
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr5_-_19988293
|
3.006
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr20_+_18794369
|
3.003
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr15_-_52587851
|
2.968
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr16_+_2880172
|
2.913
|
NM_145252
|
ZG16B
|
zymogen granule protein 16 homolog B (rat)
|
|
chr22_-_30867944
|
2.906
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr12_+_53817638
|
2.893
|
NM_001164690
NM_001164691
NM_020547
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr16_+_776957
|
2.852
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr17_+_4981721
|
2.745
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr16_+_57481336
|
2.719
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr21_-_35828038
|
2.718
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr9_+_126773851
|
2.710
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr17_+_45908992
|
2.704
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr5_+_176875325
|
2.704
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr18_-_12377168
|
2.648
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr1_-_217263195
|
2.639
|
NM_001243505
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr9_-_130639939
|
2.604
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr22_-_36019363
|
2.603
|
NM_203377
|
MB
|
myoglobin
|
|
chrX_-_112066353
|
2.592
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr20_+_31823801
|
2.576
|
NM_001243193
NM_016583
NM_130852
|
BPIFA1
|
BPI fold containing family A, member 1
|
|
chr6_-_97345760
|
2.574
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr3_-_122512526
|
2.557
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr1_+_54359859
|
2.544
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr11_-_119076888
|
2.529
|
|
CCDC153
|
coiled-coil domain containing 153
|
|
chr1_-_217262946
|
2.526
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_-_42811959
|
2.502
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr19_-_7293876
|
2.485
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr11_+_17332454
|
2.452
|
|
|
|
|
chr16_-_28621279
|
2.423
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chrX_-_112084006
|
2.408
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr11_+_73661363
|
2.403
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr3_+_160473995
|
2.402
|
NM_139245
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L
|
|
chr3_+_108015392
|
2.384
|
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr15_+_43809805
|
2.374
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr12_-_91505215
|
2.370
|
|
LUM
|
lumican
|
|
chr12_+_6898705
|
2.369
|
|
CD4
|
CD4 molecule
|
|
chr11_+_61248584
|
2.365
|
NM_001170753
NM_145017
|
PPP1R32
|
protein phosphatase 1, regulatory subunit 32
|
|
chr16_+_28875181
|
2.363
|
NM_001145812
NM_015503
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr15_+_74218794
|
2.360
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr6_+_27833033
|
2.330
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr16_-_54319966
|
2.327
|
|
IRX3
|
iroquois homeobox 3
|
|
chr4_+_77610610
|
2.323
|
|
SHROOM3
|
shroom family member 3
|
|
chr3_-_151102536
|
2.311
|
NM_022788
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr15_+_74218786
|
2.310
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr13_-_52585472
|
2.292
|
NM_000053
NM_001005918
NM_001243182
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr13_+_31506809
|
2.226
|
NM_152325
|
C13orf26
|
chromosome 13 open reading frame 26
|
|
chr2_-_31030271
|
2.221
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr1_-_27998700
|
2.183
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr4_+_41700736
|
2.170
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_52774460
|
2.163
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr11_+_63706433
|
2.153
|
NM_024771
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr15_+_74218811
|
2.134
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr16_-_16317290
|
2.113
|
NM_001079528
NM_001171
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr5_-_16617085
|
2.109
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr2_+_63277936
|
2.109
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_-_27998658
|
2.103
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr20_+_58179601
|
2.103
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_-_5248180
|
2.094
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr11_-_62472901
|
2.081
|
|
|
|
|
chr17_+_55334373
|
2.079
|
NM_170721
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr22_+_41865112
|
2.073
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr20_-_3149070
|
2.070
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr11_+_1940798
|
2.064
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr22_-_24110054
|
2.048
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr4_+_72102249
|
2.042
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr12_+_6898605
|
2.038
|
NM_000616
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr17_-_46035109
|
2.023
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr22_+_32149962
|
2.014
|
|
DEPDC5
|
DEP domain containing 5
|
|
chr1_+_67218104
|
2.007
|
NM_152665
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr20_+_32077780
|
2.002
|
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr3_-_45267068
|
1.987
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr14_+_24674902
|
1.981
|
NM_001184739
NM_174944
|
TSSK4
|
testis-specific serine kinase 4
|
|
chr1_+_162467622
|
1.978
|
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr10_-_123353771
|
1.972
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_-_27998719
|
1.971
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr16_+_2588000
|
1.962
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr15_-_93632161
|
1.948
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr3_+_133464883
|
1.947
|
NM_001063
|
TF
|
transferrin
|
|
chr5_+_76506705
|
1.947
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr15_+_74218962
|
1.936
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr5_+_121465214
|
1.932
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr15_+_59730272
|
1.917
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr2_-_73460326
|
1.917
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chr16_+_2588021
|
1.916
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr8_+_136469557
|
1.912
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr8_+_110346549
|
1.909
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr8_-_110704019
|
1.903
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr3_+_69985750
|
1.902
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr22_-_36013303
|
1.901
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr15_-_93631751
|
1.901
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr20_+_31755882
|
1.894
|
NM_080574
|
BPIFA2
|
BPI fold containing family A, member 2
|
|
chr16_+_28875077
|
1.889
|
NM_001145795
NM_001145796
NM_001145797
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr9_-_3525973
|
1.883
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr5_+_50679239
|
1.879
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr10_+_114133838
|
1.878
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr5_+_32173481
|
1.871
|
|
LOC153546
|
uncharacterized LOC153546
|
|
chr10_-_50122279
|
1.868
|
NM_001006939
|
LRRC18
|
leucine rich repeat containing 18
|
|
chr14_-_106641585
|
1.863
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_31366529
|
1.861
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr12_+_109577201
|
1.860
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr18_-_12377248
|
1.854
|
NM_006796
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr16_-_12897693
|
1.846
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr9_-_138391696
|
1.844
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr16_+_2588051
|
1.829
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr5_-_139943829
|
1.826
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr10_-_123353330
|
1.820
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_+_68165660
|
1.816
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_-_37034609
|
1.813
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr9_-_3525721
|
1.794
|
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr13_-_86373328
|
1.789
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr9_-_90589593
|
1.783
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr17_-_38074842
|
1.780
|
NM_001042471
NM_001165958
NM_001165959
|
GSDMB
|
gasdermin B
|
|
chr5_-_160279045
|
1.774
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr3_-_9439122
|
1.758
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr9_-_90589360
|
1.757
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr3_+_133465250
|
1.750
|
|
TF
|
transferrin
|
|
chr3_-_58652469
|
1.731
|
NM_138805
|
FAM3D
|
family with sequence similarity 3, member D
|
|
chr1_+_162467635
|
1.729
|
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr11_-_86383355
|
1.722
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr21_-_35883570
|
1.714
|
NM_000219
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr19_-_11545900
|
1.714
|
NM_145045
|
CCDC151
|
coiled-coil domain containing 151
|
|
chr22_+_32150002
|
1.708
|
NM_001007188
NM_001242896
NM_014662
|
DEPDC5
|
DEP domain containing 5
|
|
chr16_+_1386197
|
1.708
|
NM_001199096
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr3_-_187454247
|
1.707
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr20_+_54933982
|
1.703
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr2_+_98330030
|
1.699
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr16_+_2587989
|
1.688
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr15_-_42186274
|
1.685
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr3_+_133465204
|
1.680
|
|
TF
|
transferrin
|
|
chr20_-_25038501
|
1.678
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr6_-_47253962
|
1.665
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr2_+_121103670
|
1.649
|
NM_002193
|
INHBB
|
inhibin, beta B
|