|
chr3_+_63638343
|
6.411
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr1_-_183622447
|
3.689
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr14_-_106926393
|
3.607
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_117160748
|
2.855
|
|
AKNA
|
AT-hook transcription factor
|
|
chr2_-_239148545
|
2.650
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr9_-_75567915
|
2.645
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr2_-_239148626
|
2.603
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr9_-_75568232
|
2.586
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr18_-_53089722
|
2.574
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr17_+_72732958
|
2.442
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr6_-_52668599
|
2.409
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr6_+_32407618
|
2.368
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr11_+_27062508
|
2.343
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr1_-_149858114
|
2.258
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chrX_-_48776282
|
2.138
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr1_-_228645517
|
2.065
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr6_+_26124372
|
1.950
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr19_+_41430123
|
1.942
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr6_-_32908583
|
1.941
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr16_+_3019341
|
1.876
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr6_+_26217145
|
1.850
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr11_+_537521
|
1.845
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr9_-_3223007
|
1.727
|
|
|
|
|
chr16_+_3019466
|
1.694
|
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr6_-_27860927
|
1.652
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr14_-_106774311
|
1.594
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr9_+_136325086
|
1.539
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr13_-_36429798
|
1.472
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr3_+_186648273
|
1.471
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr5_+_36608430
|
1.403
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr18_+_44526786
|
1.363
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr6_+_27833033
|
1.353
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr10_-_123356158
|
1.351
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chrX_-_24665454
|
1.348
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr6_-_170599568
|
1.282
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr1_+_18807423
|
1.250
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr1_-_149814264
|
1.225
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr19_-_44124013
|
1.216
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr1_+_61547625
|
1.203
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_149822627
|
1.193
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr6_-_26216871
|
1.190
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr19_-_46088021
|
1.185
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr6_-_26124130
|
1.182
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr15_+_84115979
|
1.169
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr17_-_73775822
|
1.148
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr1_-_162838550
|
1.146
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chrX_-_117119282
|
1.135
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr8_+_56792345
|
1.119
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_+_149858466
|
1.107
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr19_-_44123732
|
1.103
|
|
ZNF428
|
zinc finger protein 428
|
|
chr6_+_27114860
|
1.102
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr8_+_56792392
|
1.097
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr6_-_27100528
|
1.090
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr8_+_56792399
|
1.030
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr2_-_211035913
|
1.029
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr7_+_90338711
|
1.015
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr14_+_105953256
|
1.010
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr20_+_9198036
|
1.003
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr11_-_5248180
|
0.995
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr22_+_22735202
|
0.989
|
|
|
|
|
chr10_+_106113513
|
0.955
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr19_+_41305041
|
0.930
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr13_-_61989209
|
0.928
|
|
PCDH20
|
protocadherin 20
|
|
chr19_+_41305100
|
0.925
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr3_-_64211111
|
0.908
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_201979689
|
0.908
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_+_65007418
|
0.906
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_+_97426571
|
0.901
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr1_-_149859465
|
0.893
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr2_-_97405799
|
0.877
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr17_+_14204326
|
0.876
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr6_+_27100784
|
0.875
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr19_+_17337012
|
0.867
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr15_-_64995388
|
0.846
|
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr13_+_97794050
|
0.841
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_-_39564857
|
0.834
|
NM_001144033
NM_145286
|
STOML3
|
stomatin (EPB72)-like 3
|
|
chr6_-_143266283
|
0.799
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr11_+_112047088
|
0.792
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr1_-_6052481
|
0.792
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr4_-_170077807
|
0.785
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr6_+_26158332
|
0.772
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr14_-_106967528
|
0.771
|
|
IGHA1
IGHA2
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr5_-_73936299
|
0.750
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_+_120107348
|
0.750
|
NM_001244682
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr15_+_84116172
|
0.745
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr1_-_6052410
|
0.732
|
|
NPHP4
|
nephronophthisis 4
|
|
chr8_-_33370552
|
0.717
|
NM_025115
NM_001102401
|
TTI2
|
TELO2 interacting protein 2
|
|
chr5_+_94727047
|
0.714
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr16_+_85645014
|
0.699
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr2_-_97405775
|
0.698
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chrX_+_72667089
|
0.697
|
NM_005193
|
CDX4
|
caudal type homeobox 4
|
|
chr6_+_148663728
|
0.695
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr4_-_18023358
|
0.694
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr14_+_65007163
|
0.689
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr19_+_1269244
|
0.676
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr6_-_168479796
|
0.664
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr6_+_148663953
|
0.662
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_167298382
|
0.658
|
|
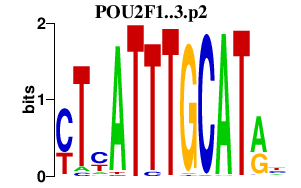
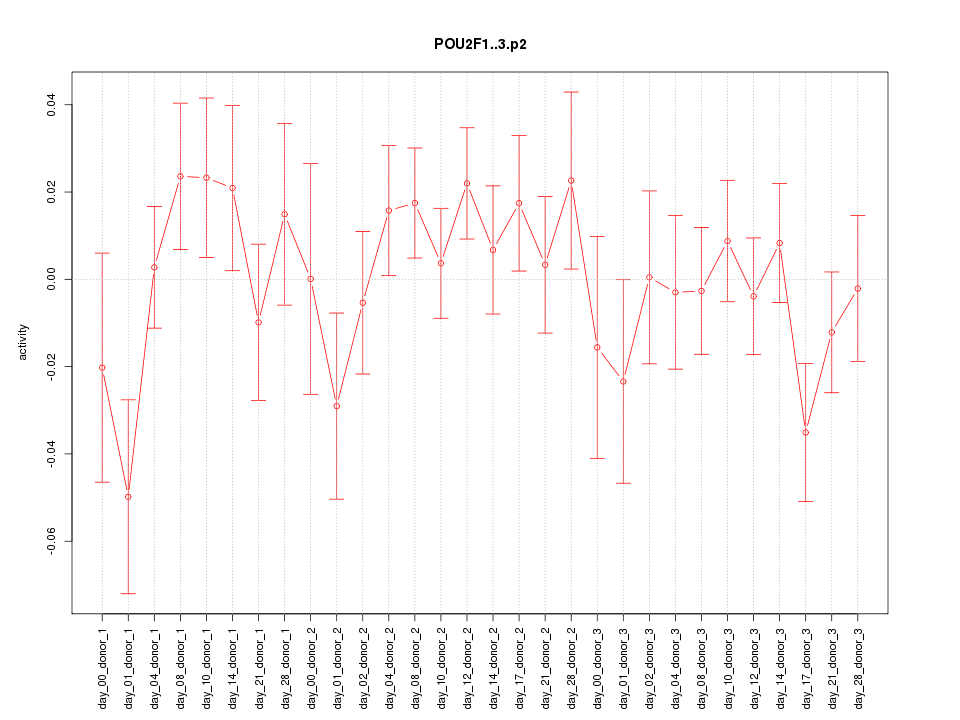
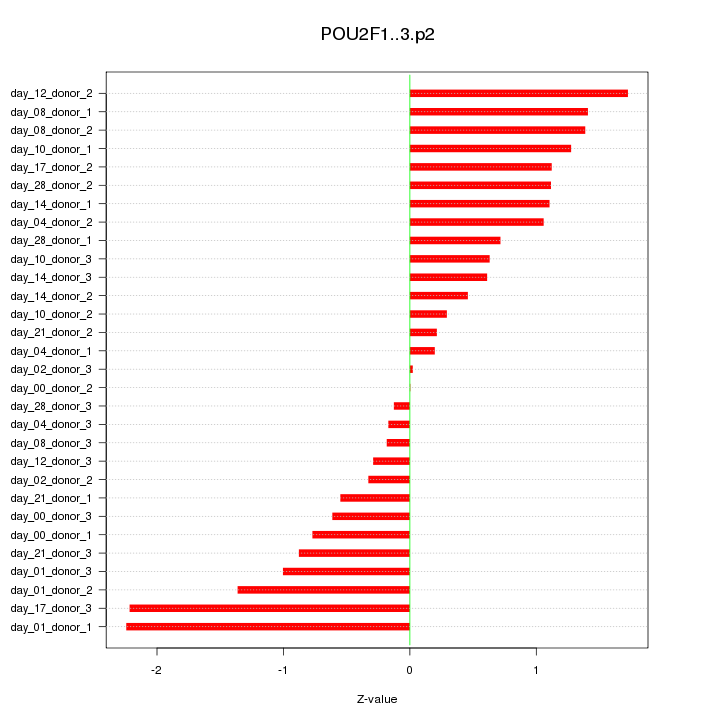
POU2F1
|
POU class 2 homeobox 1
|
|
chr3_-_129118241
|
0.643
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr1_+_61547388
|
0.636
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_201979747
|
0.629
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr9_-_32526164
|
0.621
|
NM_014314
|
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
|
|
chr19_-_46088074
|
0.616
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr7_-_112727832
|
0.615
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr17_-_19651586
|
0.594
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr16_+_53164784
|
0.593
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr15_-_64995459
|
0.590
|
NM_002537
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr19_+_41305333
|
0.586
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr1_-_149399159
|
0.576
|
NM_001024599
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr12_+_122064448
|
0.567
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr12_+_7053180
|
0.548
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chrX_+_154611748
|
0.547
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chrX_-_154688260
|
0.547
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chr18_-_52988916
|
0.546
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr1_-_94586678
|
0.543
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr2_-_239197091
|
0.536
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr7_-_112727799
|
0.529
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr6_-_138820580
|
0.528
|
|
|
|
|
chr1_+_228645807
|
0.524
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr15_-_41166336
|
0.524
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr14_-_107099379
|
0.510
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr7_-_112727778
|
0.506
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr22_-_43010894
|
0.504
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr8_-_72268720
|
0.503
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_-_57443858
|
0.501
|
NM_005379
|
MYO1A
|
myosin IA
|
|
chr12_-_31479084
|
0.497
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr17_+_57642885
|
0.497
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr8_-_72274466
|
0.491
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr17_+_63133501
|
0.490
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr3_+_119316694
|
0.489
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr19_-_19144304
|
0.488
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr19_-_19144262
|
0.488
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr14_-_106641585
|
0.487
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_-_62369213
|
0.478
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr17_+_38498270
|
0.477
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr3_+_186648515
|
0.475
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr6_+_27215479
|
0.472
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr11_-_62369250
|
0.471
|
NM_004739
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr6_-_168476531
|
0.471
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr11_-_62369097
|
0.471
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr22_-_43010942
|
0.466
|
NM_032311
NM_178136
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr19_-_46088084
|
0.463
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr5_+_73109342
|
0.462
|
NM_001244364
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr22_-_43010837
|
0.459
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr19_-_19144326
|
0.456
|
NM_001017392
NM_014884
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr1_-_149783910
|
0.448
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr12_-_31479038
|
0.446
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_-_31479120
|
0.445
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr7_-_141673572
|
0.444
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr17_+_72733350
|
0.436
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr11_-_62358969
|
0.422
|
NM_022830
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific
|
|
chr6_-_11111955
|
0.420
|
NM_207582
|
ERVFRD-1
|
endogenous retrovirus group FRD, member 1
|
|
chr15_-_64995346
|
0.420
|
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr5_-_131879172
|
0.420
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chrX_-_154114431
|
0.417
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr22_-_43010892
|
0.415
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr3_+_132036276
|
0.411
|
|
ACPP
|
acid phosphatase, prostate
|
|
chr13_-_70682458
|
0.409
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr6_+_170599790
|
0.408
|
|
FAM120B
|
family with sequence similarity 120B
|
|
chr7_+_6793739
|
0.407
|
NM_173565
NM_001099697
|
RSPH10B
RSPH10B2
|
radial spoke head 10 homolog B (Chlamydomonas)
radial spoke head 10 homolog B2 (Chlamydomonas)
|
|
chr5_-_145895675
|
0.406
|
NM_194251
|
GPR151
|
G protein-coupled receptor 151
|
|
chr1_+_167298280
|
0.406
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr8_+_86099909
|
0.406
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr11_+_112046207
|
0.406
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr8_+_19796840
|
0.404
|
|
LPL
|
lipoprotein lipase
|
|
chr9_+_109686436
|
0.401
|
|
ZNF462
|
zinc finger protein 462
|
|
chr22_+_23040394
|
0.400
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr14_-_106610338
|
0.399
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_-_129872719
|
0.399
|
NM_020228
NM_199437
|
PRDM10
|
PR domain containing 10
|
|
chr13_-_20110884
|
0.398
|
NM_001141968
NM_130785
NM_199254
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2
|
|
chr1_+_59775749
|
0.398
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr11_-_27494232
|
0.396
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr14_-_107066061
|
0.393
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_69726751
|
0.388
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr6_-_55740358
|
0.386
|
NM_021073
|
BMP5
|
bone morphogenetic protein 5
|
|
chr3_+_132036210
|
0.385
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr11_+_34642651
|
0.375
|
|
EHF
|
ets homologous factor
|
|
chr6_+_143999058
|
0.374
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_-_56359794
|
0.371
|
NM_001200053
NM_001200054
|
PMEL
|
premelanosome protein
|
|
chrX_+_69674914
|
0.370
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr8_-_26724748
|
0.369
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr10_-_64576123
|
0.369
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr8_+_42396297
|
0.367
|
NM_001135676
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr5_+_76114832
|
0.363
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr10_-_62332666
|
0.363
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_-_40596781
|
0.356
|
NM_001010880
NM_001142577
NM_001142578
NM_001142579
|
ZNF780A
|
zinc finger protein 780A
|
|
chr15_-_89089792
|
0.354
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chrX_-_80457440
|
0.343
|
NM_030763
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr6_-_35656610
|
0.342
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr11_+_34642587
|
0.342
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr17_-_36105005
|
0.342
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr3_+_113251217
|
0.339
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr22_-_36556781
|
0.337
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr20_-_40246991
|
0.337
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr7_+_27778991
|
0.335
|
NM_001206901
|
TAX1BP1
|
Tax1 (human T-cell leukemia virus type I) binding protein 1
|
|
chr7_+_94539209
|
0.333
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr2_+_86668270
|
0.333
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr9_+_129677046
|
0.332
|
NM_001190729
NM_001190730
NM_014636
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr4_-_74088773
|
0.332
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr21_+_30400229
|
0.332
|
|
USP16
|
ubiquitin specific peptidase 16
|