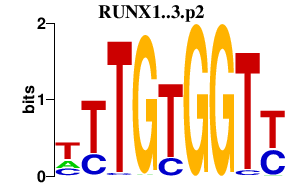
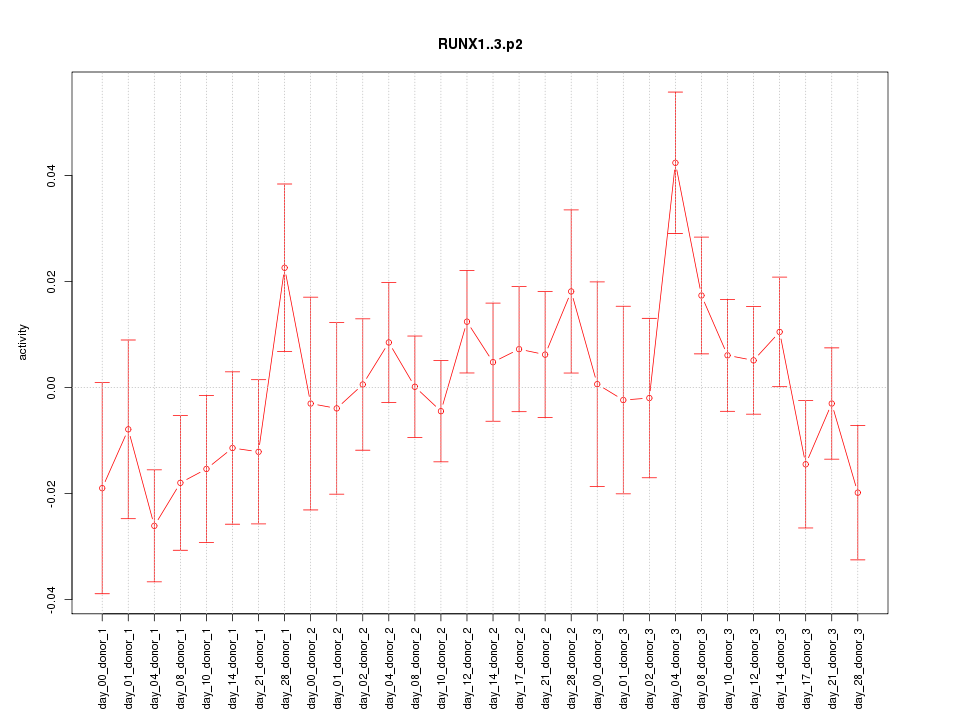
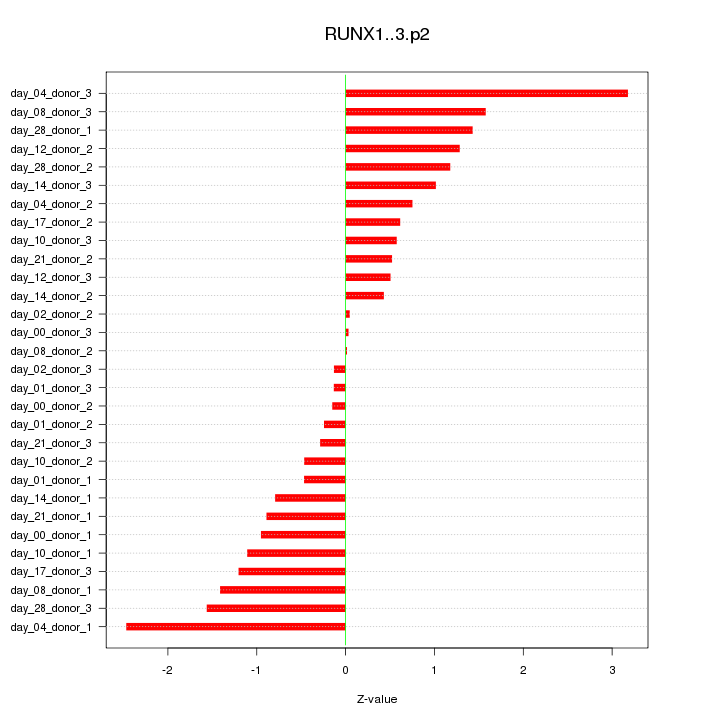
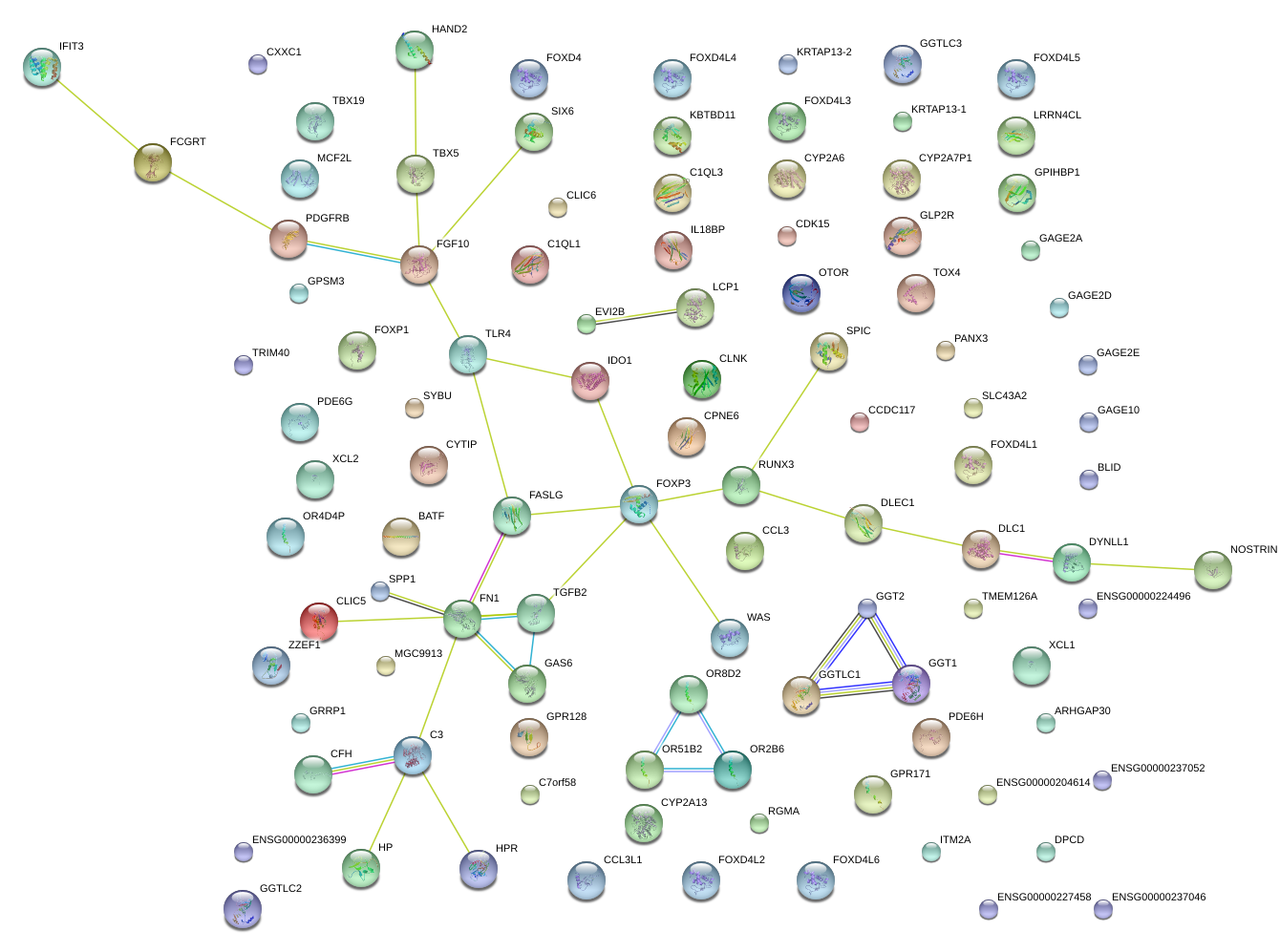
|
chr17_-_29641068
|
2.421
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr2_-_216300770
|
1.933
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr20_-_23967431
|
1.874
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr3_+_100328432
|
1.841
|
NM_032787
|
GPR128
|
G protein-coupled receptor 128
|
|
chr7_+_37723378
|
1.528
|
|
|
|
|
chr8_+_39771327
|
1.432
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr22_+_22988781
|
1.326
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr3_-_150920946
|
1.320
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr17_+_56232493
|
1.280
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chr3_-_71179736
|
1.073
|
|
FOXP1
|
forkhead box P1
|
|
chr19_+_56989482
|
1.060
|
|
LOC100128252
|
uncharacterized LOC100128252
|
|
chr17_+_9745847
|
1.016
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr12_+_15125955
|
0.994
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr17_-_29641094
|
0.972
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr2_-_216300444
|
0.953
|
|
FN1
|
fibronectin 1
|
|
chr19_-_41356193
|
0.953
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr13_+_113741668
|
0.923
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr2_-_216241321
|
0.898
|
|
FN1
|
fibronectin 1
|
|
chr14_+_24540742
|
0.884
|
NM_006032
|
CPNE6
|
copine VI (neuronal)
|
|
chr15_-_93631751
|
0.860
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93632161
|
0.859
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_196621007
|
0.854
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr14_+_75988765
|
0.820
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr14_+_75988838
|
0.794
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr6_+_30104509
|
0.750
|
NM_138700
|
TRIM40
|
tripartite motif containing 40
|
|
chr9_+_120466607
|
0.746
|
|
TLR4
|
toll-like receptor 4
|
|
chr13_-_114538953
|
0.744
|
NM_001143945
|
GAS6
|
growth arrest-specific 6
|
|
chr4_-_174450529
|
0.741
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr6_-_32163299
|
0.740
|
NM_022107
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_+_172628157
|
0.731
|
NM_000639
|
FASLG
|
Fas ligand (TNF superfamily, member 6)
|
|
chr19_-_6720585
|
0.705
|
NM_000064
|
C3
|
complement component 3
|
|
chr8_+_144295067
|
0.683
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr5_-_44388666
|
0.671
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr14_+_60975937
|
0.665
|
NM_007374
|
SIX6
|
SIX homeobox 6
|
|
chr17_-_34524054
|
0.665
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr5_-_149535420
|
0.664
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr20_+_16728997
|
0.661
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr9_-_70429730
|
0.654
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr9_-_70178814
|
0.636
|
NM_001126334
|
FOXD4L5
|
forkhead box D4-like 5
|
|
chr11_-_124190092
|
0.629
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr9_+_42717233
|
0.628
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr1_-_25291474
|
0.627
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chrX_-_78623006
|
0.625
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr11_-_5345530
|
0.623
|
NM_033180
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2
|
|
chr12_-_114846191
|
0.617
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr6_-_46047978
|
0.612
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr2_+_169659099
|
0.611
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr4_-_10686314
|
0.609
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr7_+_120628867
|
0.591
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr1_-_168513201
|
0.589
|
NM_003175
|
XCL1
XCL2
|
chemokine (C motif) ligand 1
chemokine (C motif) ligand 2
|
|
chr1_+_26485510
|
0.585
|
NM_024869
|
FAM110D
|
family with sequence similarity 110, member D
|
|
chr4_+_88896868
|
0.583
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_218518675
|
0.581
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr11_-_62457001
|
0.566
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_161039586
|
0.560
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr7_+_120628722
|
0.559
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr1_-_161039649
|
0.547
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_+_202671197
|
0.545
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr17_-_43045612
|
0.544
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chrX_-_49121171
|
0.539
|
NM_001114377
NM_014009
|
FOXP3
|
forkhead box P3
|
|
chr21_+_31768294
|
0.538
|
NM_181599
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chrX_+_48542159
|
0.533
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr11_+_124481451
|
0.530
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr19_+_50015535
|
0.528
|
NM_001136019
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr19_+_56989549
|
0.525
|
|
LOC100128252
|
uncharacterized LOC100128252
|
|
chr2_-_158300517
|
0.524
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr4_+_88896899
|
0.522
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_-_110620220
|
0.520
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_+_71710971
|
0.519
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr10_+_91092238
|
0.514
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr17_-_34417386
|
0.514
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr13_-_46756295
|
0.514
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr8_+_1952875
|
0.514
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr17_-_29641087
|
0.513
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr22_+_25003610
|
0.510
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr16_+_72088507
|
0.509
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr11_-_121986800
|
0.506
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr17_-_1532109
|
0.506
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr4_+_88896801
|
0.505
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_-_161039703
|
0.503
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chrX_+_49160124
|
0.499
|
NM_001098413
|
GAGE10
|
G antigen 10
|
|
chr4_+_88896838
|
0.497
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_32930638
|
0.495
|
NM_001145720
|
ZBTB8B
|
zinc finger and BTB domain containing 8B
|
|
chr19_+_41430123
|
0.493
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr11_-_58345593
|
0.478
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr7_+_27135672
|
0.470
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr1_+_156211950
|
0.453
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr10_+_114154522
|
0.450
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_+_50015868
|
0.449
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr12_-_58026846
|
0.449
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr1_-_161039745
|
0.447
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr17_+_80317122
|
0.445
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr19_+_51226638
|
0.444
|
|
CLEC11A
|
C-type lectin domain family 11, member A
|
|
chr6_-_32013806
|
0.443
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr2_+_128175989
|
0.441
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr17_-_73840495
|
0.440
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr3_-_45267068
|
0.437
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr17_-_39183453
|
0.434
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr6_-_46889620
|
0.433
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr5_-_137071462
|
0.423
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr20_+_54987167
|
0.420
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr18_+_61575203
|
0.419
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr3_+_133495925
|
0.416
|
|
TF
|
transferrin
|
|
chr8_+_24151579
|
0.410
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chrX_-_112066353
|
0.410
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr11_+_71710662
|
0.407
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr8_-_60031545
|
0.407
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr19_+_41497182
|
0.404
|
NM_000767
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr1_-_161039413
|
0.394
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr10_-_105238996
|
0.393
|
NM_001129742
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr8_-_12973752
|
0.391
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_+_150690838
|
0.389
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr12_+_54410641
|
0.387
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr6_+_31543340
|
0.387
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr17_-_1531543
|
0.387
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr4_+_89299874
|
0.381
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr1_+_20617411
|
0.380
|
NM_001039500
|
VWA5B1
|
von Willebrand factor A domain containing 5B1
|
|
chr11_+_124095343
|
0.380
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr11_-_10715297
|
0.380
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr4_+_8951476
|
0.376
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr4_+_646965
|
0.375
|
NM_001145292
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta
|
|
chr5_-_180018465
|
0.372
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr7_+_100183926
|
0.371
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr1_+_159796478
|
0.370
|
NM_020125
|
SLAMF8
|
SLAM family member 8
|
|
chr1_-_184006491
|
0.368
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chrX_+_119005733
|
0.367
|
NM_004541
|
NDUFA1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa
|
|
chr17_+_1633754
|
0.367
|
|
WDR81
|
WD repeat domain 81
|
|
chr17_-_56606710
|
0.366
|
NM_004574
|
SEPT4
|
septin 4
|
|
chr1_+_156339002
|
0.366
|
NM_020407
|
RHBG
|
Rh family, B glycoprotein (gene/pseudogene)
|
|
chr5_-_76934508
|
0.362
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr17_+_32582295
|
0.361
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr3_-_147124595
|
0.361
|
NM_001243256
NM_032153
|
ZIC4
|
Zic family member 4
|
|
chr6_+_25279765
|
0.358
|
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr6_-_134498973
|
0.356
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_71781091
|
0.355
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr11_-_62457370
|
0.355
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_11188800
|
0.354
|
|
|
|
|
chr5_+_147258273
|
0.353
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr17_+_47288068
|
0.351
|
|
ABI3
|
ABI family, member 3
|
|
chr3_+_15643451
|
0.351
|
|
BTD
|
biotinidase
|
|
chr12_-_14849427
|
0.349
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr8_+_85095702
|
0.348
|
|
RALYL
|
RALY RNA binding protein-like
|
|
chr1_+_22303417
|
0.347
|
NM_007352
|
CELA3A
CELA3B
|
chymotrypsin-like elastase family, member 3A
chymotrypsin-like elastase family, member 3B
|
|
chr1_+_152850797
|
0.347
|
NM_030663
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr6_-_52109226
|
0.346
|
NM_052872
|
IL17F
|
interleukin 17F
|
|
chr5_+_140174443
|
0.344
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr2_-_29297126
|
0.344
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr19_+_50796820
|
0.342
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr2_-_217724781
|
0.341
|
NM_003284
|
TNP1
|
transition protein 1 (during histone to protamine replacement)
|
|
chr17_+_40274755
|
0.340
|
NM_033194
|
HSPB9
|
heat shock protein, alpha-crystallin-related, B9
|
|
chr5_+_156696351
|
0.338
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr6_+_31539875
|
0.337
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr3_-_40350948
|
0.336
|
|
FLJ33065
|
uncharacterized LOC440952
|
|
chr2_-_234652623
|
0.331
|
NM_001001394
|
DNAJB3
|
DnaJ (Hsp40) homolog, subfamily B, member 3
|
|
chr19_+_15052298
|
0.330
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chrX_-_128788913
|
0.330
|
NM_017413
|
APLN
|
apelin
|
|
chr7_+_2552162
|
0.328
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr12_-_4754275
|
0.328
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr6_-_31846751
|
0.327
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr16_+_53133063
|
0.326
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr11_+_76745434
|
0.325
|
NM_138706
|
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase)
|
|
chr15_-_26961911
|
0.323
|
NM_001191320
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr15_-_50558161
|
0.322
|
NM_002112
|
HDC
|
histidine decarboxylase
|
|
chr5_-_176326314
|
0.320
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr22_-_37975937
|
0.320
|
NM_006498
|
LGALS2
|
lectin, galactoside-binding, soluble, 2
|
|
chr16_-_1922161
|
0.318
|
NM_001163560
|
C16orf73
|
chromosome 16 open reading frame 73
|
|
chr12_-_54121203
|
0.317
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr2_-_74756823
|
0.317
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr1_+_84864214
|
0.316
|
NM_021233
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr7_+_150745604
|
0.314
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr17_+_44076615
|
0.311
|
NM_001007532
|
STH
|
saitohin
|
|
chr15_+_50647105
|
0.310
|
|
LOC100129387
|
uncharacterized LOC100129387
|
|
chr2_+_128176019
|
0.310
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chrX_-_145896248
|
0.308
|
NM_001144064
NM_001244892
|
CXorf51A
CXorf51B
|
chromosome X open reading frame 51A
chromosome X open reading frame 51B
|
|
chr5_+_35856990
|
0.305
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr6_-_31846766
|
0.304
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr12_+_13349716
|
0.302
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr20_-_45985411
|
0.301
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr5_-_108063960
|
0.301
|
|
HP07349
|
uncharacterized protein HP07349
|
|
chr9_-_69202203
|
0.301
|
NM_001085476
|
FOXD4L6
|
forkhead box D4-like 6
|
|
chr19_-_54761057
|
0.299
|
NM_001081442
NM_001081443
NM_006840
|
LILRB5
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5
|
|
chr6_+_25279651
|
0.299
|
NM_001173977
NM_017640
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr19_-_49567123
|
0.298
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr17_-_10276321
|
0.298
|
NM_003802
|
MYH13
|
myosin, heavy chain 13, skeletal muscle
|
|
chr6_+_31540092
|
0.297
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr8_+_21900389
|
0.297
|
NM_003867
|
FGF17
|
fibroblast growth factor 17
|
|
chrX_+_145891301
|
0.294
|
NM_001144064
NM_001244892
|
CXorf51A
CXorf51B
|
chromosome X open reading frame 51A
chromosome X open reading frame 51B
|
|
chr2_-_74756815
|
0.291
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr12_-_8693391
|
0.290
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr4_-_76928601
|
0.289
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr3_-_10547267
|
0.288
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr8_-_146078355
|
0.287
|
|
COMMD5
|
COMM domain containing 5
|
|
chr22_+_31003069
|
0.287
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr2_+_204732509
|
0.285
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr9_+_120466625
|
0.284
|
|
TLR4
|
toll-like receptor 4
|
|
chr3_-_187454247
|
0.283
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr21_-_36421586
|
0.283
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_+_67222817
|
0.278
|
NM_145200
|
CABP4
|
calcium binding protein 4
|
|
chr6_-_30525370
|
0.277
|
NM_005275
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr3_+_25470067
|
0.277
|
|
RARB
|
retinoic acid receptor, beta
|